[English] 日本語
 Yorodumi
Yorodumi- EMDB-0134: Structural snapshots of the Type 9 protein translocon Plug-complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0134 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural snapshots of the Type 9 protein translocon Plug-complex | |||||||||
 Map data Map data | SprA-PPI-Plug complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type 9 Secretion System Type IX Secretion System T9S folded protein secretion outer membrane protein / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) | |||||||||
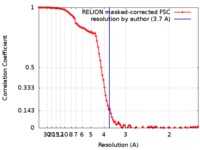
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Deme JC / Lea SM | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Type 9 secretion system structures reveal a new protein transport mechanism. Authors: Frédéric Lauber / Justin C Deme / Susan M Lea / Ben C Berks /  Abstract: The type 9 secretion system (T9SS) is the protein export pathway of bacteria of the Gram-negative Fibrobacteres-Chlorobi-Bacteroidetes superphylum and is an essential determinant of pathogenicity in ...The type 9 secretion system (T9SS) is the protein export pathway of bacteria of the Gram-negative Fibrobacteres-Chlorobi-Bacteroidetes superphylum and is an essential determinant of pathogenicity in severe periodontal disease. The central element of the T9SS is a so-far uncharacterized protein-conducting translocon located in the bacterial outer membrane. Here, using cryo-electron microscopy, we provide structural evidence that the translocon is the T9SS protein SprA. SprA forms an extremely large (36-strand) single polypeptide transmembrane β-barrel. The barrel pore is capped on the extracellular end, but has a lateral opening to the external membrane surface. Structures of SprA bound to different components of the T9SS show that partner proteins control access to the lateral opening and to the periplasmic end of the pore. Our results identify a protein transporter with a distinctive architecture that uses an alternating access mechanism in which the two ends of the protein-conducting channel are open at different times. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0134.map.gz emd_0134.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0134-v30.xml emd-0134-v30.xml emd-0134.xml emd-0134.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0134_fsc.xml emd_0134_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0134.png emd_0134.png | 61.5 KB | ||
| Filedesc metadata |  emd-0134.cif.gz emd-0134.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0134 http://ftp.pdbj.org/pub/emdb/structures/EMD-0134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0134 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0134 | HTTPS FTP |
-Validation report
| Summary document |  emd_0134_validation.pdf.gz emd_0134_validation.pdf.gz | 318.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0134_full_validation.pdf.gz emd_0134_full_validation.pdf.gz | 318.1 KB | Display | |
| Data in XML |  emd_0134_validation.xml.gz emd_0134_validation.xml.gz | 11.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0134 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0134 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0134 | HTTPS FTP |
-Related structure data
| Related structure data |  6h3jMC  0133C  6h3iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0134.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0134.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SprA-PPI-Plug complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of SprA, PPI and Plug
| Entire | Name: Complex of SprA, PPI and Plug |
|---|---|
| Components |
|
-Supramolecule #1: Complex of SprA, PPI and Plug
| Supramolecule | Name: Complex of SprA, PPI and Plug / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Molecular weight | Theoretical: 335 KDa |
-Macromolecule #1: Protein involved in gliding motility SprA
| Macromolecule | Name: Protein involved in gliding motility SprA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Molecular weight | Theoretical: 270.251719 KDa |
| Sequence | String: MRKICIFLLV LFCGNVLRSQ VKPAVQDTTK TQFSVGKMEL ENPPSILSAY KYDPITDRYI YTTSVDGFSI DYPLVLTPKE YEDLLLKES RRDYFRKKMD AIDGKKTGAE AAKKDLLPRY YINSSLFESI FGSNTIDVKP TGSVEMDLGV RYTKQDNPAF S PRNRSSLT ...String: MRKICIFLLV LFCGNVLRSQ VKPAVQDTTK TQFSVGKMEL ENPPSILSAY KYDPITDRYI YTTSVDGFSI DYPLVLTPKE YEDLLLKES RRDYFRKKMD AIDGKKTGAE AAKKDLLPRY YINSSLFESI FGSNTIDVKP TGSVEMDLGV RYTKQDNPAF S PRNRSSLT FDFDQRISMS LMGKIGTRLE VNANYDTQST FAFQNLFKLA YTPSEDDIIQ KVEVGNVSMP LNSTLIRGAQ SL FGVKTQL QFGRTTITGV FSEQKSQTKS VVAENGGTVQ NFDLYALDYD NDRHFFLSQY FRNKYDVSLK NYPFIDSRVQ ITR LEVWVT NKQNRVTTTG GGNNLRNIIA LQDLGEAQVS GVPDNEVVVI SSTAGFFNNP IDSPTSNTNN KYDPATIGQA GSFL NSNIR EIVTAKSGFN NTNVSEATDY SVLENARKLT TNEYTFNPQL GYISLQQRLA NDEILAVAFE YTVGGKVYQV GEFGS DGVD ATVVTGNNSS NQAIITQSLV LKMLKSNLTN VKNPVWNLMM KNVYQIPQAY QIKQDDFRLN ILYTDPSPIN YITPVQ GSS FPPNPAPDSK VEQTPLLNVF NLDRLNYNND PQAGGDGFFD YIPGVTVDVQ NGRVIFTTKE PFGELIFNKL QTGAGES YN DPTTYNANQQ KYVFRNMYRN TQAGALQDSD KNKFLLRGKY KSSGSNGIPI GAFNVPQGSV VVTAAGRVLV EGIDYSVD Y QLGRVQILDP SLQASNTPIE VSLENNSIFG QQTRRFMGFN IEHKISDKFV IGGTYLKMTE RPFTQKSTYG QESVNNTIF GFNGNYSTEV PFLTRLANKL PNIDTDVPSN LSIRGEVAFL RPDAPKASDF QGEATIYVDD FEGSQSTIDM RSAYAWSLAS TPFITSIND NTFNANSNTL EYGFKRAKLS WYTIDPVFYS SKPSGISNDD LSLNTTRRIY SRELYPNTDI AQGQIQVVNT L DLTYYPGE RGPYNNNPSF GASNPSANFG GIMRALNSTN FEQGNVEYIQ FWVLDPYVGN GESPATNAGK IYFNLGEISE DV LKDGRKQ YENGLGPDQV MVNPQPLWGD VPASQSLIYA FDTNPDNRKN QDVGLDGLPS SREGSIYTNY AGEADPTGDD YTY YLNADG GVLERYKNYN GTEGNSAVSI NDPNRGSTTL PDVEDINRDN TMSTINAYYE YSIDVKPGMQ VGENYITDIR EVTN VDLPN GGTTNARWIQ FKIPVSQPQN TIGNITDFRS IRFMRMFMTG FNSQMTVRFG ALDLVRGEWR RYTGTLDAND QNPDD DGVE FDVAAVNIQE NGTKCPVNYV MPPGVQREQL YNNNTVINQN EQALAVRIGG AGLQYQDSRA VFKNVSVDMR QYKKLK MFL HAESLPNQPT LEDDEMVGFI RFGNDFTQNF YQVEIPLKVT KTGGSCSISP DLVWMDDNSI DLALDLLTRM KIKAMSI DI NSSKRDVNGI YYPDNDPDLE GGDGDGKLTL GIKGNPNFGL VRNLMVGVKS RADHKDIKGE VWFNELRLAD LENKGGMA A ILNVDTNMAD FATVSATGRK STIGFGSLEQ GANERDREDV QQYNIVTNLN LGKLLPKKWG INLPFNYAIG EEVITPEYD PFNQDIKLDQ LIRETTDQAE KDNIRTRAID YTKRKSINFI GVRKDRAPEQ KPHVYDIENF TFSQSYNQVE RHDYEVADYE DEQSNSAVN YAYTFQPKEV VPFKSTKFMK KSEYWKLLSD FNFNYLPSNI SFNTNILRQS NRQQFREVEV EGIGLDPLYR R NFAFNYQY GFGFNLTKSL KLNYSATSNN IVRNFLNDDN SPKEDFNIWD DYLDIGTPNQ HAQQLVLNYD IPINKIPIFG FV KASYSYT ADYMWQRSST AFSEYEDPNG TVYDLGNTIQ NSNSNTLTTT LNMNTLYKYL GLTPGAKKTA KPKTAAPPKP GEK IVNTAK PVVSSSPFYD GLIGVLTSIK NVQINYTKNS GTVLPGYTPS VGFLGTSKPS LGFVFGSQDD VRYEAAKRGW LTTY QDFNQ SFTQVSNKLL KVTANIDLLP DLKVDLSMDR SYSENTSEQY SVDPSTNEYK PLSPYTYGMF SISTVMIKTA FSPSD ETQS AAFDDFRSNR LIIANRLAEG HYGSGVAIPR YGDANNPIPA ETDPNYAVYT ANQGYPIGYT KSNQAVLLPA FLAAYT GSD ASSSSTNIFR SFPIPNWSIK YNGLMRYKYF KDKFKRFSLQ HNYRASYTIN QFRSNFDYNS SPKVQDVNTN FYNEIIM SN VNLVEQFSPL IRMDFELKSS LRVLSEIKKD RALSMSFDNN LLTEVKGMEY IIGLGYRFKD VIFSSRLADN PTGIIKSD I NIKADFSLRN NETLVRYLDY DNNQLAAGQN IWSLKLTADY SFSKNLTAIF YYDHSFSKAV ISTSFPLTNI RSGFTLRYN FGN UniProtKB: Protein involved in gliding motility SprA |
-Macromolecule #2: Peptidyl-prolyl cis-trans isomerase
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) / Strain: ATCC 17061 / DSM 2064 / UW101 Flavobacterium johnsoniae (bacteria) / Strain: ATCC 17061 / DSM 2064 / UW101 |
| Molecular weight | Theoretical: 19.219141 KDa |
| Sequence | String: MKQLLTALLS LTLFISCSKD KDEVKDYTAE NEKEIVDYLA QNNLTAQRTN SGLYYIITKE GSSESEGENP GEEENTGEGE NTEENENDG HPTLNSNITV IYKGYFTNGK VFDESTEGVS YSLRTLIPGW KEGIPLLKSG GEIQLFVPAH LGYGSNGNKT V PGGAVLIF EITLVSVN UniProtKB: Peptidyl-prolyl cis-trans isomerase |
-Macromolecule #3: Plug
| Macromolecule | Name: Plug / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Molecular weight | Theoretical: 48.602969 KDa |
| Sequence | String: MPKFLYQSLI ALLIFTSAKA QEIQTEVVPP YNIKTVTFVQ NGNNVVPIFE LNSTFEFQFD DLFGNEANYY FEITHCDYNW KPSDIPKTD YLRGFDGQRI MDYSNSFNTL QVYSHYRLPF PNQFTTQIRL SGNYILKILN EDKEVVLSRK FIVYEEHCTV G AQVKRTRN ...String: MPKFLYQSLI ALLIFTSAKA QEIQTEVVPP YNIKTVTFVQ NGNNVVPIFE LNSTFEFQFD DLFGNEANYY FEITHCDYNW KPSDIPKTD YLRGFDGQRI MDYSNSFNTL QVYSHYRLPF PNQFTTQIRL SGNYILKILN EDKEVVLSRK FIVYEEHCTV G AQVKRTRN LSNINYKQNL DFTIASNDIT FQTPTQNVKV LLLQNGNFNT AIKNIPPQYT IGNQLVYKYD AETQFWGGNE FL YFENKDI RAANNNVGRI GSNNDIYNAY LYTNAARGNQ IYTNYQDVNG NFVVKNINGA DNSIEADYAW VYFTLSAPAF RMN KDIYIT GIFNNYSLSP EYKMDYNTDK AVFEKAVMIK QGFTNYQYTV ADKKGNIDLE NAIDGNFYQT ENEYTILVYY KESI DRYQR VIGKGNANSI NIVN UniProtKB: Type 9 secretion system plug protein N-terminal domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: Correlation |
|---|---|
| Output model |  PDB-6h3j: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)