+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ste | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NKCC1 Fu_CTD | ||||||
 Components Components | Solute carrier family 12 member 2 | ||||||
 Keywords Keywords |  TRANSPORT PROTEIN / cotransporter / dimer / TRANSPORT PROTEIN / cotransporter / dimer /  ion / ion /  membrane protein membrane protein | ||||||
| Function / homology |  Function and homology information Function and homology informationchloride:monoatomic cation symporter activity / sodium ion transport /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.34 Å cryo EM / Resolution: 3.34 Å | ||||||
 Authors Authors | Moseng, M.A. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling. Authors: Mitchell A Moseng / Chih-Chia Su / Kerri Rios / Meng Cui / Meinan Lyu / Przemyslaw Glaza / Philip A Klenotic / Eric Delpire / Edward W Yu /  Abstract: The Na-K-2Cl cotransporter-1 (NKCC1) is an electroneutral Na-dependent transporter responsible for simultaneously translocating Na, K, and Cl ions into cells. In human tissue, NKCC1 plays a critical ...The Na-K-2Cl cotransporter-1 (NKCC1) is an electroneutral Na-dependent transporter responsible for simultaneously translocating Na, K, and Cl ions into cells. In human tissue, NKCC1 plays a critical role in regulating cytoplasmic volume, fluid intake, chloride homeostasis, and cell polarity. Here, we report four structures of human NKCC1 (hNKCC1), both in the absence and presence of loop diuretic (bumetanide or furosemide), using single-particle cryo-electron microscopy. These structures allow us to directly observe various novel conformations of the hNKCC1 dimer. They also reveal two drug-binding sites located at the transmembrane and cytosolic carboxyl-terminal domains, respectively. Together, our findings enable us to delineate an inhibition mechanism that involves a coupled movement between the cytosolic and transmembrane domains of hNKCC1. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ste.cif.gz 8ste.cif.gz | 133.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ste.ent.gz pdb8ste.ent.gz | 107.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ste.json.gz 8ste.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/st/8ste https://data.pdbj.org/pub/pdb/validation_reports/st/8ste ftp://data.pdbj.org/pub/pdb/validation_reports/st/8ste ftp://data.pdbj.org/pub/pdb/validation_reports/st/8ste | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  40759MC  7mxoC  7n3nC  7sflC  7smpC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
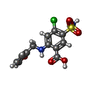
| #1: Protein | Mass: 45190.438 Da / Num. of mol.: 2 / Fragment: NKCC1 Fu_CTD Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: SLC12A2 / Production host: Homo sapiens (human) / Gene: SLC12A2 / Production host:   Homo sapiens (human) / References: UniProt: A0A8C9GPS0 Homo sapiens (human) / References: UniProt: A0A8C9GPS0#2: Chemical |  Furosemide FurosemideHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: NKCC1_Fu_CTD / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 5000 nm / Nominal defocus min: 1200 nm Bright-field microscopy / Nominal defocus max: 5000 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 40 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING ONLY |
|---|---|
3D reconstruction | Resolution: 3.34 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 304440 / Symmetry type: POINT |
 Movie
Movie Controller
Controller



 PDBj
PDBj