[English] 日本語
 Yorodumi
Yorodumi- PDB-8fu3: Structure Of Respiratory Syncytial Virus Polymerase with Novel No... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8fu3 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  VIRAL PROTEIN / RSV / VIRAL PROTEIN / RSV /  Polymerase / Non-Nucleoside Inhibitor / Polymerase / Non-Nucleoside Inhibitor /  TRANSFERASE TRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication /  Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides /  viral life cycle / viral life cycle /  virion component / virion component /  : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm /  RNA-directed RNA polymerase ...NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / RNA-directed RNA polymerase ...NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication /  Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides /  viral life cycle / viral life cycle /  virion component / virion component /  : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm /  RNA-directed RNA polymerase / RNA-directed RNA polymerase /  RNA-dependent RNA polymerase activity / RNA-dependent RNA polymerase activity /  GTPase activity / GTPase activity /  ATP binding / ATP binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||
| Biological species |   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.88 Å cryo EM / Resolution: 2.88 Å | ||||||
 Authors Authors | Yu, X. / Abeywickrema, P. / Bonneux, B. / Behera, I. / Jacoby, E. / Fung, A. / Adhikary, S. / Bhaumik, A. / Carbajo, R.J. / Bruyn, S.D. ...Yu, X. / Abeywickrema, P. / Bonneux, B. / Behera, I. / Jacoby, E. / Fung, A. / Adhikary, S. / Bhaumik, A. / Carbajo, R.J. / Bruyn, S.D. / Miller, R. / Patrick, A. / Pham, Q. / Piassek, M. / Verheyen, N. / Shareef, A. / Sutto-Ortiz, P. / Ysebaert, N. / Vlijmen, H.V. / Jonckers, T.H.M. / Herschke, F. / McLellan, J.S. / Decroly, E. / Fearns, R. / Grosse, S. / Roymans, D. / Sharma, S. / Rigaux, P. / Jin, Z. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor. Authors: Xiaodi Yu / Pravien Abeywickrema / Brecht Bonneux / Ishani Behera / Brandon Anson / Edgar Jacoby / Amy Fung / Suraj Adhikary / Anusarka Bhaumik / Rodrigo J Carbajo / Suzanne De Bruyn / Robyn ...Authors: Xiaodi Yu / Pravien Abeywickrema / Brecht Bonneux / Ishani Behera / Brandon Anson / Edgar Jacoby / Amy Fung / Suraj Adhikary / Anusarka Bhaumik / Rodrigo J Carbajo / Suzanne De Bruyn / Robyn Miller / Aaron Patrick / Quyen Pham / Madison Piassek / Nick Verheyen / Afzaal Shareef / Priscila Sutto-Ortiz / Nina Ysebaert / Herman Van Vlijmen / Tim H M Jonckers / Florence Herschke / Jason S McLellan / Etienne Decroly / Rachel Fearns / Sandrine Grosse / Dirk Roymans / Sujata Sharma / Peter Rigaux / Zhinan Jin /     Abstract: The respiratory syncytial virus polymerase complex, consisting of the polymerase (L) and phosphoprotein (P), catalyzes nucleotide polymerization, cap addition, and cap methylation via the RNA ...The respiratory syncytial virus polymerase complex, consisting of the polymerase (L) and phosphoprotein (P), catalyzes nucleotide polymerization, cap addition, and cap methylation via the RNA dependent RNA polymerase, capping, and Methyltransferase domains on L. Several nucleoside and non-nucleoside inhibitors have been reported to inhibit this polymerase complex, but the structural details of the exact inhibitor-polymerase interactions have been lacking. Here, we report a non-nucleoside inhibitor JNJ-8003 with sub-nanomolar inhibition potency in both antiviral and polymerase assays. Our 2.9 Å resolution cryo-EM structure revealed that JNJ-8003 binds to an induced-fit pocket on the capping domain, with multiple interactions consistent with its tight binding and resistance mutation profile. The minigenome and gel-based de novo RNA synthesis and primer extension assays demonstrated that JNJ-8003 inhibited nucleotide polymerization at the early stages of RNA transcription and replication. Our results support that JNJ-8003 binding modulates a functional interplay between the capping and RdRp domains, and this molecular insight could accelerate the design of broad-spectrum antiviral drugs. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8fu3.cif.gz 8fu3.cif.gz | 375.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8fu3.ent.gz pdb8fu3.ent.gz | 282.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8fu3.json.gz 8fu3.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fu/8fu3 https://data.pdbj.org/pub/pdb/validation_reports/fu/8fu3 ftp://data.pdbj.org/pub/pdb/validation_reports/fu/8fu3 ftp://data.pdbj.org/pub/pdb/validation_reports/fu/8fu3 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29452MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 254482.750 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human respiratory syncytial virus A2 / Production host: Human respiratory syncytial virus A2 / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm)References: UniProt: P28887,  RNA-directed RNA polymerase, mRNA (guanine-N7)-methyltransferase, RNA-directed RNA polymerase, mRNA (guanine-N7)-methyltransferase,  Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, GDP polyribonucleotidyltransferase Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases, GDP polyribonucleotidyltransferase | ||||
|---|---|---|---|---|---|
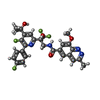
| #2: Protein |  / Protein P / Protein PMass: 29062.895 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human respiratory syncytial virus A2 / Production host: Human respiratory syncytial virus A2 / Production host:   Spodoptera frugiperda (fall armyworm) / References: UniProt: P03421 Spodoptera frugiperda (fall armyworm) / References: UniProt: P03421#3: Chemical | ChemComp-YBK / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Source (recombinant) | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2400 nm / Nominal defocus min: 1200 nm Bright-field microscopy / Nominal defocus max: 2400 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 52 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
3D reconstruction | Resolution: 2.88 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 519118 / Symmetry type: POINT |
 Movie
Movie Controller
Controller


 PDBj
PDBj