[English] 日本語
 Yorodumi
Yorodumi- EMDB-29452: Structure Of Respiratory Syncytial Virus Polymerase with Novel No... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RSV /  Polymerase / Non-Nucleoside Inhibitor / Polymerase / Non-Nucleoside Inhibitor /  VIRAL PROTEIN / VIRAL PROTEIN /  TRANSFERASE TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication /  Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides /  viral life cycle / viral life cycle /  virion component / virion component /  : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm /  RNA-directed RNA polymerase ...NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / RNA-directed RNA polymerase ...NNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication /  Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides /  viral life cycle / viral life cycle /  virion component / virion component /  : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm / : / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / host cell cytoplasm /  RNA-directed RNA polymerase / RNA-directed RNA polymerase /  RNA-dependent RNA polymerase activity / RNA-dependent RNA polymerase activity /  GTPase activity / GTPase activity /  ATP binding / ATP binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.88 Å cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Yu X / Abeywickrema P / Bonneux B / Behera I / Jacoby E / Fung A / Adhikary S / Bhaumik A / Carbajo RJ / Bruyn SD ...Yu X / Abeywickrema P / Bonneux B / Behera I / Jacoby E / Fung A / Adhikary S / Bhaumik A / Carbajo RJ / Bruyn SD / Miller R / Patrick A / Pham Q / Piassek M / Verheyen N / Shareef A / Sutto-Ortiz P / Ysebaert N / Vlijmen HV / Jonckers THM / Herschke F / McLellan JS / Decroly E / Fearns R / Grosse S / Roymans D / Sharma S / Rigaux P / Jin Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Structural and mechanistic insights into the inhibition of respiratory syncytial virus polymerase by a non-nucleoside inhibitor. Authors: Xiaodi Yu / Pravien Abeywickrema / Brecht Bonneux / Ishani Behera / Brandon Anson / Edgar Jacoby / Amy Fung / Suraj Adhikary / Anusarka Bhaumik / Rodrigo J Carbajo / Suzanne De Bruyn / Robyn ...Authors: Xiaodi Yu / Pravien Abeywickrema / Brecht Bonneux / Ishani Behera / Brandon Anson / Edgar Jacoby / Amy Fung / Suraj Adhikary / Anusarka Bhaumik / Rodrigo J Carbajo / Suzanne De Bruyn / Robyn Miller / Aaron Patrick / Quyen Pham / Madison Piassek / Nick Verheyen / Afzaal Shareef / Priscila Sutto-Ortiz / Nina Ysebaert / Herman Van Vlijmen / Tim H M Jonckers / Florence Herschke / Jason S McLellan / Etienne Decroly / Rachel Fearns / Sandrine Grosse / Dirk Roymans / Sujata Sharma / Peter Rigaux / Zhinan Jin /     Abstract: The respiratory syncytial virus polymerase complex, consisting of the polymerase (L) and phosphoprotein (P), catalyzes nucleotide polymerization, cap addition, and cap methylation via the RNA ...The respiratory syncytial virus polymerase complex, consisting of the polymerase (L) and phosphoprotein (P), catalyzes nucleotide polymerization, cap addition, and cap methylation via the RNA dependent RNA polymerase, capping, and Methyltransferase domains on L. Several nucleoside and non-nucleoside inhibitors have been reported to inhibit this polymerase complex, but the structural details of the exact inhibitor-polymerase interactions have been lacking. Here, we report a non-nucleoside inhibitor JNJ-8003 with sub-nanomolar inhibition potency in both antiviral and polymerase assays. Our 2.9 Å resolution cryo-EM structure revealed that JNJ-8003 binds to an induced-fit pocket on the capping domain, with multiple interactions consistent with its tight binding and resistance mutation profile. The minigenome and gel-based de novo RNA synthesis and primer extension assays demonstrated that JNJ-8003 inhibited nucleotide polymerization at the early stages of RNA transcription and replication. Our results support that JNJ-8003 binding modulates a functional interplay between the capping and RdRp domains, and this molecular insight could accelerate the design of broad-spectrum antiviral drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29452.map.gz emd_29452.map.gz | 78.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29452-v30.xml emd-29452-v30.xml emd-29452.xml emd-29452.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29452_fsc.xml emd_29452_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_29452.png emd_29452.png | 37.8 KB | ||
| Filedesc metadata |  emd-29452.cif.gz emd-29452.cif.gz | 7.4 KB | ||
| Others |  emd_29452_half_map_1.map.gz emd_29452_half_map_1.map.gz emd_29452_half_map_2.map.gz emd_29452_half_map_2.map.gz | 65.4 MB 65.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29452 http://ftp.pdbj.org/pub/emdb/structures/EMD-29452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29452 | HTTPS FTP |
-Related structure data
| Related structure data |  8fu3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29452.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29452.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29452_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
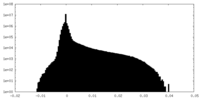
| Projections & Slices |
| ||||||||||||
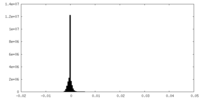
| Density Histograms |
-Half map: #2
| File | emd_29452_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
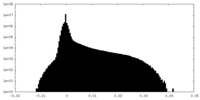
| Projections & Slices |
| ||||||||||||
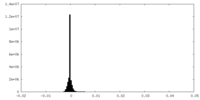
| Density Histograms |
- Sample components
Sample components
-Entire : Structure Of Respiratory Syncytial Virus Polymerase with Novel No...
| Entire | Name: Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: Structure Of Respiratory Syncytial Virus Polymerase with Novel No...
| Supramolecule | Name: Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  RNA-directed RNA polymerase RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 254.48275 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MGSWSHPQFE KGSGSGSSWS HPQFEKGSGS LVPRGSMDPI INGNSANVYL TDSYLKGVIS FSECNALGSY IFNGPYLKND YTNLISRQN PLIEHMNLKK LNITQSLISK YHKGEIKLEE PTYFQSLLMT YKSMTSSEQI ATTNLLKKII RRAIEISDVK V YAILNKLG ...String: MGSWSHPQFE KGSGSGSSWS HPQFEKGSGS LVPRGSMDPI INGNSANVYL TDSYLKGVIS FSECNALGSY IFNGPYLKND YTNLISRQN PLIEHMNLKK LNITQSLISK YHKGEIKLEE PTYFQSLLMT YKSMTSSEQI ATTNLLKKII RRAIEISDVK V YAILNKLG LKEKDKIKSN NGQDEDNSVI TTIIKDDILS AVKDNQSHLK ADKNHSTKQK DTIKTTLLKK LMCSMQHPPS WL IHWFNLY TKLNNILTQY RSNEVKNHGF TLIDNQTLSG FQFILNQYGC IVYHKELKRI TVTTYNQFLT WKDISLSRLN VCL ITWISN CLNTLNKSLG LRCGFNNVIL TQLFLYGDCI LKLFHNEGFY IIKEVEGFIM SLILNITEED QFRKRFYNSM LNNI TDAAN KAQKNLLSRV CHTLLDKTVS DNIINGRWII LLSKFLKLIK LAGDNNLNNL SELYFLFRIF GHPMVDERQA MDAVK INCN ETKFYLLSSL SMLRGAFIYR IIKGFVNNYN RWPTLRNAIV LPLRWLTYYK LNTYPSLLEL TERDLIVLSG LRFYRE FRL PKKVDLEMII NDKAISPPKN LIWTSFPRNY MPSHIQNYIE HEKLKFSESD KSRRVLEYYL RDNKFNECDL YNCVVNQ SY LNNPNHVVSL TGKERELSVG RMFAMQPGMF RQVQILAEKM IAENILQFFP ESLTRYGDLE LQKILELKAG ISNKSNRY N DNYNNYISKC SIITDLSKFN QAFRYETSCI CSDVLDELHG VQSLFSWLHL TIPHVTIICT YRHAPPYIGD HIVDLNNVD EQSGLYRYHM GGIEGWCQKL WTIEAISLLD LISLKGKFSI TALINGDNQS IDISKPIRLM EGQTHAQADY LLALNSLKLL YKEYAGIGH KLKGTETYIS RDMQFMSKTI QHNGVYYPAS IKKVLRVGPW INTILDDFKV SLESIGSLTQ ELEYRGESLL C SLIFRNVW LYNQIALQLK NHALCNNKLY LDILKVLKHL KTFFNLDNID TALTLYMNLP MLFGGGDPNL LYRSFYRRTP DF LTEAIVH SVFILSYYTN HDLKDKLQDL SDDRLNKFLT CIITFDKNPN AEFVTLMRDP QALGSERQAK ITSEINRLAV TEV LSTAPN KIFSKSAQHY TTTEIDLNDI MQNIEPTYPH GLRVVYESLP FYKAEKIVNL ISGTKSITNI LEKTSAIDLT DIDR ATEMM RKNITLLIRI LPLDCNRDKR EILSMENLSI TELSKYVRER SWSLSNIVGV TSPSIMYTMD IKYTTSTISS GIIIE KYNV NSLTRGERGP TKPWVGSSTQ EKKTMPVYNR QVLTKKQRDQ IDLLAKLDWV YASIDNKDEF MEELSIGTLG LTYEKA KKL FPQYLSVNYL HRLTVSSRPC EFPASIPAYR TTNYHFDTSP INRILTEKYG DEDIDIVFQN CISFGLSLMS VVEQFTN VC PNRIILIPKL NEIHLMKPPI FTGDVDIHKL KQVIQKQHMF LPDKISLTQY VELFLSNKTL KSGSHVNSNL ILAHKISD Y FHNTYILSTN LAGHWILIIQ LMKDSKGIFE KDWGEGYITD HMFINLKVFF NAYKTYLLCF HKGYGKAKLE CDMNTSDLL CVLELIDSSY WKSMSKVFLE QKVIKYILSQ DASLHRVKGC HSFKLWFLKR LNVAEFTVCP WVVNIDYHPT HMKAILTYID LVRMGLINI DRIHIKNKHK FNDEFYTSNL FYINYNFSDN THLLTKHIRI ANSELENNYN KLYHPTPETL ENILANPIKS N DKKTLNDY CIGKNVDSIM LPLLSNKKLI KSSAMIRTNY SKQDLYNLFP MVVIDRIIDH SGNTAKSNQL YTTTSHQISL VH NSTSLYC MLPWHHINRF NFVFSSTGCK ISIEYILKDL KIKDPNCIAF IGEGAGNLLL RTVVELHPDI RYIYRSLKDC NDH SLPIEF LRLYNGHINI DYGENLTIPA TDATNNIHWS YLHIKFAEPI SLFVCDAELS VTVNWSKIII EWSKHVRKCK YCSS VNKCM LIVKYHAQDD IDFKLDNITI LKTYVCLGSK LKGSEVYLVL TIGPANIFPV FNVVQNAKLI LSRTKNFIMP KKADK ESID ANIKSLIPFL CYPITKKGIN TALSKLKSVV SGDILSYSIA GRNEVFSNKL INHKHMNILK WFNHVLNFRS TELNYN HLY MVESTYPYLS ELLNSLTTNE LKKLIKITGS LLYNFHNE UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 29.062895 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD ...String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD AMIGLREEMI EKIRTEALMT NDRLEAMARL RNEESEKMAK DTSDEVSLNP TSEKLNNLLE GNDSDNDLSL ED FKGENKY FQGHHHHHH UniProtKB:  Phosphoprotein Phosphoprotein |
-Macromolecule #3: 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluor...
| Macromolecule | Name: 8-methoxy-3-methyl-N-{(2S)-3,3,3-trifluoro-2-[5-fluoro-6-(4-fluorophenyl)-4-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-hydroxypropyl}cinnoline-6-carboxamide type: ligand / ID: 3 / Number of copies: 1 / Formula: YBK |
|---|---|
| Molecular weight | Theoretical: 576.515 Da |
| Chemical component information | 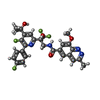 ChemComp-YBK: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X