[English] 日本語
 Yorodumi
Yorodumi- PDB-7yu4: Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-07... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7yu4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, focused on receptor | ||||||
 Components Components | Lysophosphatidic acid receptor 1 | ||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR GPCR | ||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to 1-oleoyl-sn-glycerol 3-phosphate /  lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / calmodulin dependent kinase signaling pathway / Lysosphingolipid and LPA receptors / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / calmodulin dependent kinase signaling pathway / Lysosphingolipid and LPA receptors /  lysophosphatidic acid binding / negative regulation of cilium assembly / regulation of synaptic vesicle cycle / cellular response to oxygen levels / oligodendrocyte development ...cellular response to 1-oleoyl-sn-glycerol 3-phosphate / lysophosphatidic acid binding / negative regulation of cilium assembly / regulation of synaptic vesicle cycle / cellular response to oxygen levels / oligodendrocyte development ...cellular response to 1-oleoyl-sn-glycerol 3-phosphate /  lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / calmodulin dependent kinase signaling pathway / Lysosphingolipid and LPA receptors / lysophosphatidic acid receptor activity / positive regulation of smooth muscle cell chemotaxis / calmodulin dependent kinase signaling pathway / Lysosphingolipid and LPA receptors /  lysophosphatidic acid binding / negative regulation of cilium assembly / regulation of synaptic vesicle cycle / cellular response to oxygen levels / oligodendrocyte development / corpus callosum development / bleb assembly / lysophosphatidic acid binding / negative regulation of cilium assembly / regulation of synaptic vesicle cycle / cellular response to oxygen levels / oligodendrocyte development / corpus callosum development / bleb assembly /  regulation of metabolic process / optic nerve development / negative regulation of cAMP-mediated signaling / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of dendritic spine development / positive regulation of Rho protein signal transduction / activation of phospholipase C activity / GABA-ergic synapse / G-protein alpha-subunit binding / positive regulation of stress fiber assembly / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / regulation of metabolic process / optic nerve development / negative regulation of cAMP-mediated signaling / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of dendritic spine development / positive regulation of Rho protein signal transduction / activation of phospholipase C activity / GABA-ergic synapse / G-protein alpha-subunit binding / positive regulation of stress fiber assembly / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway /  myelination / cerebellum development / myelination / cerebellum development /  neurogenesis / cell chemotaxis / dendritic shaft / neurogenesis / cell chemotaxis / dendritic shaft /  PDZ domain binding / G protein-coupled receptor activity / adenylate cyclase-activating G protein-coupled receptor signaling pathway / negative regulation of neuron projection development / PDZ domain binding / G protein-coupled receptor activity / adenylate cyclase-activating G protein-coupled receptor signaling pathway / negative regulation of neuron projection development /  presynaptic membrane / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / regulation of cell shape / G alpha (i) signalling events / presynaptic membrane / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / regulation of cell shape / G alpha (i) signalling events /  postsynaptic membrane / G alpha (q) signalling events / positive regulation of canonical NF-kappaB signal transduction / postsynaptic membrane / G alpha (q) signalling events / positive regulation of canonical NF-kappaB signal transduction /  dendritic spine / positive regulation of MAPK cascade / dendritic spine / positive regulation of MAPK cascade /  endosome / positive regulation of apoptotic process / G protein-coupled receptor signaling pathway / neuronal cell body / glutamatergic synapse / endosome / positive regulation of apoptotic process / G protein-coupled receptor signaling pathway / neuronal cell body / glutamatergic synapse /  cell surface / cell surface /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | ||||||
 Authors Authors | Akasaka, H. / Shihoya, W. / Nureki, O. | ||||||
| Funding support |  Japan, 1items Japan, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the active G-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist. Authors: Hiroaki Akasaka / Tatsuki Tanaka / Fumiya K Sano / Yuma Matsuzaki / Wataru Shihoya / Osamu Nureki /  Abstract: Lysophosphatidic acid receptor 1 (LPA) is one of the six G protein-coupled receptors activated by the bioactive lipid, lysophosphatidic acid (LPA). LPA is a drug target for various diseases, ...Lysophosphatidic acid receptor 1 (LPA) is one of the six G protein-coupled receptors activated by the bioactive lipid, lysophosphatidic acid (LPA). LPA is a drug target for various diseases, including cancer, inflammation, and neuropathic pain. Notably, LPA agonists have potential therapeutic value for obesity and urinary incontinence. Here, we report a cryo-electron microscopy structure of the active human LPA-G complex bound to ONO-0740556, an LPA analog with more potent activity against LPA. Our structure elucidated the details of the agonist binding mode and receptor activation mechanism mediated by rearrangements of transmembrane segment 7 and the central hydrophobic core. A structural comparison of LPA and other phylogenetically-related lipid-sensing GPCRs identified the structural determinants for lipid preference of LPA. Moreover, we characterized the structural polymorphisms at the receptor-G-protein interface, which potentially reflect the G-protein dissociation process. Our study provides insights into the detailed mechanism of LPA binding to agonists and paves the way toward the design of drug-like agonists targeting LPA. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7yu4.cif.gz 7yu4.cif.gz | 63.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7yu4.ent.gz pdb7yu4.ent.gz | 45.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7yu4.json.gz 7yu4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yu/7yu4 https://data.pdbj.org/pub/pdb/validation_reports/yu/7yu4 ftp://data.pdbj.org/pub/pdb/validation_reports/yu/7yu4 ftp://data.pdbj.org/pub/pdb/validation_reports/yu/7yu4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  34098MC  7yu3C  7yu5C  7yu6C  7yu7C  7yu8C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  / LPA receptor 1 / LPA-1 / Lysophosphatidic acid receptor Edg-2 / LPA receptor 1 / LPA-1 / Lysophosphatidic acid receptor Edg-2Mass: 42936.004 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: LPAR1, EDG2, LPA1 / Production host: Homo sapiens (human) / Gene: LPAR1, EDG2, LPA1 / Production host:   Spodoptera frugiperda (fall armyworm) / References: UniProt: Q92633 Spodoptera frugiperda (fall armyworm) / References: UniProt: Q92633 |
|---|---|
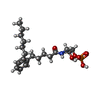
| #2: Chemical | ChemComp-K6L / [( |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human Lysophosphatidic Acid Receptor 1 bound to ONO-0740556 Type: COMPLEX / Entity ID: #1 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Buffer solution | pH: 8 |
| Specimen | Conc.: 7 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0267 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 181071 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 3.3→88.44 Å / Cor.coef. Fo:Fc: 0.803 / SU B: 32.341 / SU ML: 0.472 / ESU R: 0.592 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: PARAMETERS FOR MASK CACLULATION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 99.316 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 2251 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj