+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7yeh | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human OGT-OGA complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE/HYDROLASE /  O-GlcNAc transferase / O-GlcNAc transferase /  O-GlcNAcase / O-GlcNAcase /  Complex / Mutual inhibition / Complex / Mutual inhibition /  TRANSFERASE / TRANSFERASE-HYDROLASE complex TRANSFERASE / TRANSFERASE-HYDROLASE complex | ||||||
| Function / homology |  Function and homology information Function and homology informationglycoprotein metabolic process /  protein N-acetylglucosaminyltransferase complex / hyalurononglucosaminidase activity / protein N-acetylglucosaminyltransferase complex / hyalurononglucosaminidase activity /  protein O-GlcNAc transferase / N-acetylglucosamine metabolic process / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / glycoprotein catabolic process / protein O-GlcNAc transferase / N-acetylglucosamine metabolic process / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / glycoprotein catabolic process /  protein O-GlcNAcase ...glycoprotein metabolic process / protein O-GlcNAcase ...glycoprotein metabolic process /  protein N-acetylglucosaminyltransferase complex / hyalurononglucosaminidase activity / protein N-acetylglucosaminyltransferase complex / hyalurononglucosaminidase activity /  protein O-GlcNAc transferase / N-acetylglucosamine metabolic process / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / glycoprotein catabolic process / protein O-GlcNAc transferase / N-acetylglucosamine metabolic process / regulation of insulin receptor signaling pathway / protein O-acetylglucosaminyltransferase activity / positive regulation of transcription from RNA polymerase II promoter by glucose / glycoprotein catabolic process /  protein O-GlcNAcase / protein O-GlcNAcase /  : / : /  : / [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine/L-threonine O-N-acetyl-alpha-D-glucosaminase activity / acetylglucosaminyltransferase activity / regulation of necroptotic process / regulation of Rac protein signal transduction / negative regulation of stem cell population maintenance / protein O-linked glycosylation / protein deglycosylation / NSL complex / : / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / regulation of glycolytic process / RIPK1-mediated regulated necrosis / : / [protein]-3-O-(N-acetyl-D-glucosaminyl)-L-serine/L-threonine O-N-acetyl-alpha-D-glucosaminase activity / acetylglucosaminyltransferase activity / regulation of necroptotic process / regulation of Rac protein signal transduction / negative regulation of stem cell population maintenance / protein O-linked glycosylation / protein deglycosylation / NSL complex / : / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / regulation of glycolytic process / RIPK1-mediated regulated necrosis /  regulation of synapse assembly / regulation of synapse assembly /  regulation of gluconeogenesis / positive regulation of stem cell population maintenance / Formation of WDR5-containing histone-modifying complexes / positive regulation of proteolysis / phosphatidylinositol-3,4,5-trisphosphate binding / regulation of gluconeogenesis / positive regulation of stem cell population maintenance / Formation of WDR5-containing histone-modifying complexes / positive regulation of proteolysis / phosphatidylinositol-3,4,5-trisphosphate binding /  mitophagy / mitophagy /  hemopoiesis / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / hemopoiesis / negative regulation of proteasomal ubiquitin-dependent protein catabolic process /  histone acetyltransferase complex / positive regulation of lipid biosynthetic process / negative regulation of protein ubiquitination / positive regulation of TORC1 signaling / response to nutrient / negative regulation of cell migration / positive regulation of translation / cell projection / histone acetyltransferase complex / positive regulation of lipid biosynthetic process / negative regulation of protein ubiquitination / positive regulation of TORC1 signaling / response to nutrient / negative regulation of cell migration / positive regulation of translation / cell projection /  beta-N-acetylglucosaminidase activity / beta-N-acetylglucosaminidase activity /  mitochondrial membrane / cellular response to glucose stimulus / negative regulation of transforming growth factor beta receptor signaling pathway / circadian regulation of gene expression / response to insulin / Regulation of necroptotic cell death / protein processing / chromatin DNA binding / UCH proteinases / positive regulation of cold-induced thermogenesis / chromatin organization / HATs acetylate histones / glutamatergic synapse / apoptotic process / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / mitochondrial membrane / cellular response to glucose stimulus / negative regulation of transforming growth factor beta receptor signaling pathway / circadian regulation of gene expression / response to insulin / Regulation of necroptotic cell death / protein processing / chromatin DNA binding / UCH proteinases / positive regulation of cold-induced thermogenesis / chromatin organization / HATs acetylate histones / glutamatergic synapse / apoptotic process / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II /  signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex /  nucleoplasm / nucleoplasm /  membrane / identical protein binding / membrane / identical protein binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.92 Å cryo EM / Resolution: 3.92 Å | ||||||
 Authors Authors | Lu, P. / Liu, Y. / Yu, H. / Gao, H. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex. Authors: Ping Lu / Yusong Liu / Maozhou He / Ting Cao / Mengquan Yang / Shutao Qi / Hongtao Yu / Haishan Gao /  Abstract: O-GlcNAcylation is a conserved post-translational modification that attaches N-acetyl glucosamine (GlcNAc) to myriad cellular proteins. In response to nutritional and hormonal signals, O- ...O-GlcNAcylation is a conserved post-translational modification that attaches N-acetyl glucosamine (GlcNAc) to myriad cellular proteins. In response to nutritional and hormonal signals, O-GlcNAcylation regulates diverse cellular processes by modulating the stability, structure, and function of target proteins. Dysregulation of O-GlcNAcylation has been implicated in the pathogenesis of cancer, diabetes, and neurodegeneration. A single pair of enzymes, the O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), catalyzes the addition and removal of O-GlcNAc on over 3,000 proteins in the human proteome. However, how OGT selects its native substrates and maintains the homeostatic control of O-GlcNAcylation of so many substrates against OGA is not fully understood. Here, we present the cryo-electron microscopy (cryo-EM) structures of human OGT and the OGT-OGA complex. Our studies reveal that OGT forms a functionally important scissor-shaped dimer. Within the OGT-OGA complex structure, a long flexible OGA segment occupies the extended substrate-binding groove of OGT and positions a serine for O-GlcNAcylation, thus preventing OGT from modifying other substrates. Conversely, OGT disrupts the functional dimerization of OGA and occludes its active site, resulting in the blocking of access by other substrates. This mutual inhibition between OGT and OGA may limit the futile O-GlcNAcylation cycles and help to maintain O-GlcNAc homeostasis. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7yeh.cif.gz 7yeh.cif.gz | 453.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7yeh.ent.gz pdb7yeh.ent.gz | 351.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7yeh.json.gz 7yeh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ye/7yeh https://data.pdbj.org/pub/pdb/validation_reports/ye/7yeh ftp://data.pdbj.org/pub/pdb/validation_reports/ye/7yeh ftp://data.pdbj.org/pub/pdb/validation_reports/ye/7yeh | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 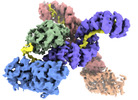 33773MC  7yeaC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 117953.414 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: OGT Homo sapiens (human) / Gene: OGTProduction host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: O15294,  protein O-GlcNAc transferase protein O-GlcNAc transferase#2: Protein |  / OGA / Beta-N-acetylglucosaminidase / Beta-N-acetylhexosaminidase / Beta-hexosaminidase / Meningioma- ...OGA / Beta-N-acetylglucosaminidase / Beta-N-acetylhexosaminidase / Beta-hexosaminidase / Meningioma-expressed antigen 5 / N-acetyl-beta-D-glucosaminidase / N-acetyl-beta-glucosaminidase / Nuclear cytoplasmic O-GlcNAcase and acetyltransferase / NCOAT / OGA / Beta-N-acetylglucosaminidase / Beta-N-acetylhexosaminidase / Beta-hexosaminidase / Meningioma- ...OGA / Beta-N-acetylglucosaminidase / Beta-N-acetylhexosaminidase / Beta-hexosaminidase / Meningioma-expressed antigen 5 / N-acetyl-beta-D-glucosaminidase / N-acetyl-beta-glucosaminidase / Nuclear cytoplasmic O-GlcNAcase and acetyltransferase / NCOATMass: 103020.906 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: OGA, HEXC, KIAA0679, MEA5, MGEA5 Homo sapiens (human) / Gene: OGA, HEXC, KIAA0679, MEA5, MGEA5Production host:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria)References: UniProt: O60502,  protein O-GlcNAcase protein O-GlcNAcase#3: Chemical | ChemComp-UDP / |  Uridine diphosphate Uridine diphosphate#4: Sugar | ChemComp-NAG / |  N-Acetylglucosamine N-AcetylglucosamineHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of human OGT-OGA complex / Type: COMPLEX / Entity ID: #1-#2 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 0.4 MDa / Experimental value: NO |
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:   Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) Escherichia coli 'BL21-Gold(DE3)pLysS AG' (bacteria) |
| Buffer solution | pH: 7.5 |
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2300 nm / Nominal defocus min: 1300 nm / Alignment procedure: COMA FREE Bright-field microscopy / Nominal defocus max: 2300 nm / Nominal defocus min: 1300 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
CTF correction | Type: NONE | ||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 114543 | ||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.92 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 114543 / Symmetry type: POINT | ||||||||||||||||||||||||
| Atomic model building | B value: 146 / Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||||||
| Atomic model building | 3D fitting-ID: 1 / Pdb chain-ID: A / Source name: PDB / Type: experimental model
| ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj














