+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7fj1 | ||||||
|---|---|---|---|---|---|---|---|
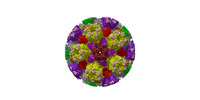
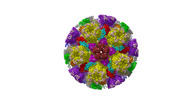
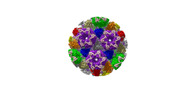
| Title | Cryo-EM structure of pseudorabies virus C-capsid | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  VIRUS / VIRUS /  Pseudorabies virus / C-capsid / Pseudorabies virus / C-capsid /  Cryo-EM Cryo-EM | ||||||
| Function / homology |  Function and homology information Function and homology informationchromosome organization => GO:0051276 / T=16 icosahedral viral capsid / deNEDDylase activity / viral genome packaging /  viral tegument / viral tegument /  viral capsid assembly / viral DNA genome replication / viral capsid assembly / viral DNA genome replication /  host cell / viral release from host cell / viral process ...chromosome organization => GO:0051276 / T=16 icosahedral viral capsid / deNEDDylase activity / viral genome packaging / host cell / viral release from host cell / viral process ...chromosome organization => GO:0051276 / T=16 icosahedral viral capsid / deNEDDylase activity / viral genome packaging /  viral tegument / viral tegument /  viral capsid assembly / viral DNA genome replication / viral capsid assembly / viral DNA genome replication /  host cell / viral release from host cell / viral process / viral penetration into host nucleus / host cell / viral release from host cell / viral process / viral penetration into host nucleus /  viral capsid / symbiont-mediated perturbation of host ubiquitin-like protein modification / host cell cytoplasm / membrane => GO:0016020 / symbiont entry into host cell / host cell nucleus / structural molecule activity / viral capsid / symbiont-mediated perturbation of host ubiquitin-like protein modification / host cell cytoplasm / membrane => GO:0016020 / symbiont entry into host cell / host cell nucleus / structural molecule activity /  DNA binding DNA bindingSimilarity search - Function | ||||||
| Biological species |    Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.43 Å cryo EM / Resolution: 4.43 Å | ||||||
 Authors Authors | Zheng, Q. / Li, S. / Zha, Z. / Sun, H. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures of pseudorabies virus capsids. Authors: Guosong Wang / Zhenghui Zha / Pengfei Huang / Hui Sun / Yang Huang / Maozhou He / Tian Chen / Lina Lin / Zhenqin Chen / Zhibo Kong / Yuqiong Que / Tingting Li / Ying Gu / Hai Yu / Jun Zhang ...Authors: Guosong Wang / Zhenghui Zha / Pengfei Huang / Hui Sun / Yang Huang / Maozhou He / Tian Chen / Lina Lin / Zhenqin Chen / Zhibo Kong / Yuqiong Que / Tingting Li / Ying Gu / Hai Yu / Jun Zhang / Qingbing Zheng / Yixin Chen / Shaowei Li / Ningshao Xia /  Abstract: Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral ...Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral encephalitis caused by PRV infection, suggesting that PRV may be able to overcome the species barrier to infect humans. To date, there is no available therapeutic for PRV infection. Here, we report the near-atomic structures of the PRV A-capsid and C-capsid, and illustrate the interaction that occurs between these subunits. We show that the C-capsid portal complex is decorated with capsid-associated tegument complexes. The PRV capsid structure is highly reminiscent of other α-herpesviruses, with some additional structural features of β- and γ-herpesviruses. These results illustrate the structure of the PRV capsid and elucidate the underlying assembly mechanism at the molecular level. This knowledge may be useful for the development of oncolytic agents or specific therapeutics against this arm of the herpesvirus family. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7fj1.cif.gz 7fj1.cif.gz | 4.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7fj1.ent.gz pdb7fj1.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7fj1.json.gz 7fj1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fj/7fj1 https://data.pdbj.org/pub/pdb/validation_reports/fj/7fj1 ftp://data.pdbj.org/pub/pdb/validation_reports/fj/7fj1 ftp://data.pdbj.org/pub/pdb/validation_reports/fj/7fj1 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  31611MC  7fj3C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | x 60
|
| 2 |
|
| 3 | x 5
|
| 4 | x 6
|
| 5 | 
|
| Symmetry | Point symmetry: (Schoenflies symbol : I (icosahedral : I (icosahedral )) )) |
- Components
Components
-Protein , 5 types, 36 molecules 0ASUaefglmnpquwyBEFGHIJKLMNOPQ...
| #1: Protein | Mass: 146101.984 Da / Num. of mol.: 16 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G8T2 Suid alphaherpesvirus 1 / References: UniProt: G3G8T2#3: Protein | | Mass: 64304.371 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G8T5 Suid alphaherpesvirus 1 / References: UniProt: G3G8T5#4: Protein | Mass: 11509.209 Da / Num. of mol.: 15 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G8R4 Suid alphaherpesvirus 1 / References: UniProt: G3G8R4#6: Protein | Mass: 57412.797 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G971 Suid alphaherpesvirus 1 / References: UniProt: G3G971#7: Protein | Mass: 7069.434 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G8Y0 Suid alphaherpesvirus 1 / References: UniProt: G3G8Y0 |
|---|
-Triplex capsid protein ... , 2 types, 15 molecules 123jkosvxzThirt
| #2: Protein | Mass: 31763.766 Da / Num. of mol.: 10 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: G3G8T3 Suid alphaherpesvirus 1 / References: UniProt: G3G8T3#5: Protein | Mass: 40008.594 Da / Num. of mol.: 5 / Source method: isolated from a natural source / Source: (natural)    Suid alphaherpesvirus 1 / References: UniProt: Q85211 Suid alphaherpesvirus 1 / References: UniProt: Q85211 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Suid alphaherpesvirus 1 Pseudorabies / Type: VIRUS / Entity ID: all / Source: NATURAL Pseudorabies / Type: VIRUS / Entity ID: all / Source: NATURAL |
|---|---|
| Source (natural) | Organism:    Suid alphaherpesvirus 1 Suid alphaherpesvirus 1 |
| Details of virus | Empty: NO / Enveloped: YES / Isolate: OTHER / Type: VIRION |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TECNAI F30 |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Electron dose: 25 e/Å2 / Film or detector model: FEI FALCON III (4k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
3D reconstruction | Resolution: 4.43 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 14252 / Symmetry type: POINT |
 Movie
Movie Controller
Controller









 PDBj
PDBj
