+Search query
-Structure paper
| Title | Structures of pseudorabies virus capsids. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 1533, Year 2022 |
| Publish date | Mar 22, 2022 |
 Authors Authors | Guosong Wang / Zhenghui Zha / Pengfei Huang / Hui Sun / Yang Huang / Maozhou He / Tian Chen / Lina Lin / Zhenqin Chen / Zhibo Kong / Yuqiong Que / Tingting Li / Ying Gu / Hai Yu / Jun Zhang / Qingbing Zheng / Yixin Chen / Shaowei Li / Ningshao Xia /  |
| PubMed Abstract | Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral ...Pseudorabies virus (PRV) is a major etiological agent of swine infectious diseases and is responsible for significant economic losses in the swine industry. Recent data points to human viral encephalitis caused by PRV infection, suggesting that PRV may be able to overcome the species barrier to infect humans. To date, there is no available therapeutic for PRV infection. Here, we report the near-atomic structures of the PRV A-capsid and C-capsid, and illustrate the interaction that occurs between these subunits. We show that the C-capsid portal complex is decorated with capsid-associated tegument complexes. The PRV capsid structure is highly reminiscent of other α-herpesviruses, with some additional structural features of β- and γ-herpesviruses. These results illustrate the structure of the PRV capsid and elucidate the underlying assembly mechanism at the molecular level. This knowledge may be useful for the development of oncolytic agents or specific therapeutics against this arm of the herpesvirus family. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35318331 / PubMed:35318331 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.31 - 6.79 Å |
| Structure data | 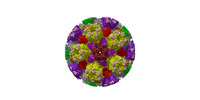 EMDB-31591: Cryo-EM structure of PRV A-capsid 5-fold sub-particle  EMDB-31592: Cryo-EM structure of PRV A-capsid 3-fold sub-particle  EMDB-31593: Cryo-EM structure of PRV A-capsid 2-fold sub-particle 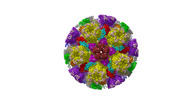 EMDB-31594: Cryo-EM structure of PRV C-capsid 5-fold sub-particle 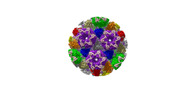 EMDB-31608: Cryo-EM structure of PRV C-capsid 3-fold sub-particle  EMDB-31609: Cryo-EM structure of PRV C-capsid 2-fold sub-particle  EMDB-31610: Cryo-EM structure of PRV C-capsid portal vertex EMDB-31611, PDB-7fj1: EMDB-31612, PDB-7fj3:  EMDB-31616: Portal-based reconstruction of PRV C-capsid |
| Source |
|
 Keywords Keywords |  VIRUS / VIRUS /  Pseudorabies virus / C-capsid / Pseudorabies virus / C-capsid /  Cryo-EM Cryo-EM |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers









