+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7c61 | ||||||
|---|---|---|---|---|---|---|---|
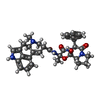
| Title | Crystal structure of 5-HT1B-BRIL and SRP2070_Fab complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  GPCR / BRIL / GPCR / BRIL /  Crystallization / Crystallization /  Antibody Antibody | ||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels / adenylate cyclase-inhibiting serotonin receptor signaling pathway / negative regulation of gamma-aminobutyric acid secretion / G protein-coupled serotonin receptor complex / serotonergic synapse /  regulation of behavior / regulation of behavior /  Serotonin receptors / negative regulation of synaptic transmission, GABAergic / response to mineralocorticoid / negative regulation of serotonin secretion ...voltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels / adenylate cyclase-inhibiting serotonin receptor signaling pathway / negative regulation of gamma-aminobutyric acid secretion / G protein-coupled serotonin receptor complex / serotonergic synapse / Serotonin receptors / negative regulation of synaptic transmission, GABAergic / response to mineralocorticoid / negative regulation of serotonin secretion ...voltage-gated calcium channel activity involved in regulation of presynaptic cytosolic calcium levels / adenylate cyclase-inhibiting serotonin receptor signaling pathway / negative regulation of gamma-aminobutyric acid secretion / G protein-coupled serotonin receptor complex / serotonergic synapse /  regulation of behavior / regulation of behavior /  Serotonin receptors / negative regulation of synaptic transmission, GABAergic / response to mineralocorticoid / negative regulation of serotonin secretion / Serotonin receptors / negative regulation of synaptic transmission, GABAergic / response to mineralocorticoid / negative regulation of serotonin secretion /  drinking behavior / cellular response to temperature stimulus / drinking behavior / cellular response to temperature stimulus /  serotonin binding / serotonin binding /  bone remodeling / negative regulation of synaptic transmission, glutamatergic / G protein-coupled serotonin receptor activity / bone remodeling / negative regulation of synaptic transmission, glutamatergic / G protein-coupled serotonin receptor activity /  vasoconstriction / protein kinase C-activating G protein-coupled receptor signaling pathway / vasoconstriction / protein kinase C-activating G protein-coupled receptor signaling pathway /  neurotransmitter receptor activity / G protein-coupled receptor internalization / regulation of synaptic vesicle exocytosis / regulation of dopamine secretion / neurotransmitter receptor activity / G protein-coupled receptor internalization / regulation of synaptic vesicle exocytosis / regulation of dopamine secretion /  heterocyclic compound binding / cellular response to alkaloid / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / heterocyclic compound binding / cellular response to alkaloid / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger /  calyx of Held / positive regulation of vascular associated smooth muscle cell proliferation / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / response to cocaine / cellular response to xenobiotic stimulus / calyx of Held / positive regulation of vascular associated smooth muscle cell proliferation / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / response to cocaine / cellular response to xenobiotic stimulus /  presynaptic membrane / G alpha (i) signalling events / chemical synaptic transmission / response to ethanol / presynaptic membrane / G alpha (i) signalling events / chemical synaptic transmission / response to ethanol /  electron transfer activity / electron transfer activity /  periplasmic space / iron ion binding / periplasmic space / iron ion binding /  dendrite / dendrite /  heme binding / heme binding /  endoplasmic reticulum / endoplasmic reticulum /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse)  Homo sapiens (human) Homo sapiens (human)  Escherichia coli (E. coli) Escherichia coli (E. coli) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 3 Å molecular replacement / Resolution: 3 Å | ||||||
 Authors Authors | Suzuki, M. / Miyagi, H. / Asada, H. / Yasunaga, M. / Suno, C. / Takahashi, Y. / Saito, J. / Iwata, S. | ||||||
 Citation Citation |  Journal: Sci Rep / Year: 2020 Journal: Sci Rep / Year: 2020Title: The discovery of a new antibody for BRIL-fused GPCR structure determination. Authors: Miyagi, H. / Asada, H. / Suzuki, M. / Takahashi, Y. / Yasunaga, M. / Suno, C. / Iwata, S. / Saito, J.I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7c61.cif.gz 7c61.cif.gz | 313.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7c61.ent.gz pdb7c61.ent.gz | 253.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7c61.json.gz 7c61.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c6/7c61 https://data.pdbj.org/pub/pdb/validation_reports/c6/7c61 ftp://data.pdbj.org/pub/pdb/validation_reports/c6/7c61 ftp://data.pdbj.org/pub/pdb/validation_reports/c6/7c61 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7c6aC  4iarS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Antibody | Mass: 23681.053 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Mus musculus (house mouse) Mus musculus (house mouse) |
|---|---|
| #2: Antibody | Mass: 35237.762 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Mus musculus (house mouse) Mus musculus (house mouse) |
| #3: Protein | Mass: 45114.523 Da / Num. of mol.: 1 / Mutation: L138W,M1007W,H1102I,R1106L Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)   Escherichia coli (E. coli) Escherichia coli (E. coli)Gene: HTR1B, HTR1DB, cybC / Production host:   Spodoptera frugiperda (fall armyworm) / References: UniProt: P28222, UniProt: P0ABE7 Spodoptera frugiperda (fall armyworm) / References: UniProt: P28222, UniProt: P0ABE7 |
| #4: Chemical | ChemComp-ERM /  Ergotamine Ergotamine |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.09 Å3/Da / Density % sol: 60.2 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 30% PEG400, 0.4M SODIUM THIOCYANATE, 0.1M SODIUM ACETATE PH 5.5 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL32XU / Wavelength: 1 Å / Beamline: BL32XU / Wavelength: 1 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Nov 6, 2015 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 3→48.5 Å / Num. obs: 25815 / % possible obs: 100 % / Redundancy: 29.5 % / CC1/2: 0.992 / Rmerge(I) obs: 0.483 / Rpim(I) all: 0.09 / Rrim(I) all: 0.491 / Net I/σ(I): 9.2 / Num. measured all: 760396 / Scaling rejects: 271 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4IAR Resolution: 3→40 Å / Cor.coef. Fo:Fc: 0.892 / Cor.coef. Fo:Fc free: 0.862 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.444 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 360.78 Å2 / Biso mean: 152.781 Å2 / Biso min: 42.31 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3→40 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3→3.078 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj