+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6kzf | ||||||
|---|---|---|---|---|---|---|---|
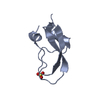
| Title | Racemic X-ray Structure of Calcicludine | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TOXIN / TOXIN /  calcicludine / racemic crystal / L-type Ca2+ channels calcicludine / racemic crystal / L-type Ca2+ channels | ||||||
| Function / homology |  Function and homology information Function and homology informationcalcium channel regulator activity / serine-type endopeptidase inhibitor activity /  toxin activity / toxin activity /  extracellular space extracellular spaceSimilarity search - Function | ||||||
| Biological species |   Dendroaspis angusticeps (eastern green mamba) Dendroaspis angusticeps (eastern green mamba) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.52 Å MOLECULAR REPLACEMENT / Resolution: 2.52 Å | ||||||
 Authors Authors | Qu, Q. / Gao, S. / Liu, L. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2020 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2020Title: Synthesis of Disulfide Surrogate Peptides Incorporating Large-Span Surrogate Bridges Through a Native-Chemical-Ligation-Assisted Diaminodiacid Strategy Authors: Qu, Q. / Gao, S. / Wu, F. / Zhang, M.G. / Li, Y. / Zhang, L.H. / Bierer, D. / Tian, C.L. / Zheng, J.S. / Liu, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6kzf.cif.gz 6kzf.cif.gz | 43.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6kzf.ent.gz pdb6kzf.ent.gz | 29.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6kzf.json.gz 6kzf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kz/6kzf https://data.pdbj.org/pub/pdb/validation_reports/kz/6kzf ftp://data.pdbj.org/pub/pdb/validation_reports/kz/6kzf ftp://data.pdbj.org/pub/pdb/validation_reports/kz/6kzf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5yv7S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6980.236 Da / Num. of mol.: 1 / Source method: obtained synthetically Source: (synth.)   Dendroaspis angusticeps (eastern green mamba) Dendroaspis angusticeps (eastern green mamba)References: UniProt: P81658 |
|---|---|
| #2: Protein | Mass: 6989.211 Da / Num. of mol.: 1 / Source method: obtained synthetically Source: (synth.)   Dendroaspis angusticeps (eastern green mamba) Dendroaspis angusticeps (eastern green mamba) |
| #3: Water | ChemComp-HOH /  Water Water |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.09 Å3/Da / Density % sol: 60.17 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop Details: 0.2M Ammonium acetate, 0.1M Sodium citrate tribasic dihydrate pH 5.6, 30% w/v Polyethylene glycol 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.54 Å |
| Detector | Type: DECTRIS PILATUS 200K / Detector: PIXEL / Date: Jun 1, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.54 Å / Relative weight: 1 : 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.52→50 Å / Num. obs: 5969 / % possible obs: 99.9 % / Redundancy: 11.4 % / Biso Wilson estimate: 36.27 Å2 / Rmerge(I) obs: 0.159 / Net I/σ(I): 14.6 |
| Reflection shell | Resolution: 2.52→2.56 Å / Rmerge(I) obs: 0.996 / Num. unique obs: 312 |
- Processing
Processing
| Software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5YV7 Resolution: 2.52→35.71 Å / Cross valid method: NONE
| ||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.52→35.71 Å
| ||||||||||||||||
| LS refinement shell | Resolution: 2.52→2.56 Å /
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




