+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6jjz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of MAGI2 and Dendrin complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  SIGNALING PROTEIN / WW tandem / PY tandem / MAGI2 / SIGNALING PROTEIN / WW tandem / PY tandem / MAGI2 /  Dendrin Dendrin | ||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of postsynaptic specialization /  extrinsic component of postsynaptic membrane / type II activin receptor binding / neuroligin clustering involved in postsynaptic membrane assembly / podocyte development / positive regulation of synaptic vesicle clustering / extrinsic component of postsynaptic membrane / type II activin receptor binding / neuroligin clustering involved in postsynaptic membrane assembly / podocyte development / positive regulation of synaptic vesicle clustering /  slit diaphragm / slit diaphragm /  beta-1 adrenergic receptor binding / structural constituent of postsynaptic density / nerve growth factor signaling pathway ...structural constituent of postsynaptic specialization / beta-1 adrenergic receptor binding / structural constituent of postsynaptic density / nerve growth factor signaling pathway ...structural constituent of postsynaptic specialization /  extrinsic component of postsynaptic membrane / type II activin receptor binding / neuroligin clustering involved in postsynaptic membrane assembly / podocyte development / positive regulation of synaptic vesicle clustering / extrinsic component of postsynaptic membrane / type II activin receptor binding / neuroligin clustering involved in postsynaptic membrane assembly / podocyte development / positive regulation of synaptic vesicle clustering /  slit diaphragm / slit diaphragm /  beta-1 adrenergic receptor binding / structural constituent of postsynaptic density / nerve growth factor signaling pathway / beta-1 adrenergic receptor binding / structural constituent of postsynaptic density / nerve growth factor signaling pathway /  activin receptor binding / activin receptor binding /  clathrin-dependent endocytosis / negative regulation of activin receptor signaling pathway / SMAD protein signal transduction / dendritic spine membrane / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / regulation of synaptic membrane adhesion / ciliary base / SMAD binding / receptor clustering / clathrin-dependent endocytosis / negative regulation of activin receptor signaling pathway / SMAD protein signal transduction / dendritic spine membrane / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / regulation of synaptic membrane adhesion / ciliary base / SMAD binding / receptor clustering /  kinesin binding / positive regulation of phosphoprotein phosphatase activity / positive regulation of receptor internalization / photoreceptor outer segment / GABA-ergic synapse / bicellular tight junction / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / kinesin binding / positive regulation of phosphoprotein phosphatase activity / positive regulation of receptor internalization / photoreceptor outer segment / GABA-ergic synapse / bicellular tight junction / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction /  phosphatase binding / phosphatase binding /  centriole / photoreceptor inner segment / negative regulation of cell migration / cellular response to nerve growth factor stimulus / cell projection / positive regulation of neuron projection development / cell-cell junction / signaling receptor complex adaptor activity / late endosome / centriole / photoreceptor inner segment / negative regulation of cell migration / cellular response to nerve growth factor stimulus / cell projection / positive regulation of neuron projection development / cell-cell junction / signaling receptor complex adaptor activity / late endosome /  postsynaptic membrane / postsynaptic membrane /  perikaryon / perikaryon /  postsynaptic density / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / negative regulation of cell population proliferation / postsynaptic density / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / negative regulation of cell population proliferation /  centrosome / glutamatergic synapse / centrosome / glutamatergic synapse /  synapse / synapse /  dendrite / protein-containing complex binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm / dendrite / protein-containing complex binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm /  signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | ||||||
 Authors Authors | Lin, Z. / Zhang, H. / Yang, Z. / Ji, Z. / Zhang, M. / Zhu, J. | ||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity. Authors: Lin, Z. / Yang, Z. / Xie, R. / Ji, Z. / Guan, K. / Zhang, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6jjz.cif.gz 6jjz.cif.gz | 106.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6jjz.ent.gz pdb6jjz.ent.gz | 80.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6jjz.json.gz 6jjz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjz https://data.pdbj.org/pub/pdb/validation_reports/jj/6jjz ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjz ftp://data.pdbj.org/pub/pdb/validation_reports/jj/6jjz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6j68C 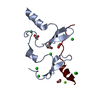 6jjwC  6jjxC  6jjyC  6jk0C  6jk1C  2ysdS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / Activin receptor-interacting protein 1 / Acvrip1 / Atrophin-1-interacting protein 1 / AIP-1 / ...Activin receptor-interacting protein 1 / Acvrip1 / Atrophin-1-interacting protein 1 / AIP-1 / Membrane-associated guanylate kinase inverted 2 / MAGI-2 / Activin receptor-interacting protein 1 / Acvrip1 / Atrophin-1-interacting protein 1 / AIP-1 / ...Activin receptor-interacting protein 1 / Acvrip1 / Atrophin-1-interacting protein 1 / AIP-1 / Membrane-associated guanylate kinase inverted 2 / MAGI-2Mass: 12089.212 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Magi2, Acvrinp1, Aip1, Arip1 / Production host: Mus musculus (house mouse) / Gene: Magi2, Acvrinp1, Aip1, Arip1 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q9WVQ1 Escherichia coli (E. coli) / References: UniProt: Q9WVQ1#2: Protein/peptide |  Mass: 1843.966 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Ddn, Gm748, Kiaa0749 / Production host: Mus musculus (house mouse) / Gene: Ddn, Gm748, Kiaa0749 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q80TS7 Escherichia coli (E. coli) / References: UniProt: Q80TS7#3: Chemical |  Acetate Acetate#4: Chemical | ChemComp-SO4 / |  Sulfate Sulfate#5: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.13 Å3/Da / Density % sol: 42.17 % |
|---|---|
Crystal grow | Temperature: 289 K / Method: vapor diffusion / pH: 4.6 Details: 2.0-2.5M ammonium sulfate, 100mM sodium acetate (pH 4.6) |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U1 / Wavelength: 0.97915 Å / Beamline: BL17U1 / Wavelength: 0.97915 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Mar 20, 2016 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97915 Å / Relative weight: 1 : 0.97915 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→50 Å / Num. obs: 29381 / % possible obs: 99.8 % / Redundancy: 6.9 % / Rmerge(I) obs: 0.047 / Net I/σ(I): 44 |
| Reflection shell | Resolution: 1.65→1.68 Å / Redundancy: 6.8 % / Rmerge(I) obs: 0.856 / Mean I/σ(I) obs: 2.5 / Num. unique obs: 1465 / % possible all: 99.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2YSD Resolution: 1.65→27.917 Å / SU ML: 0.2 / Cross valid method: FREE R-VALUE / σ(F): 1.89 / Phase error: 27.42
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→27.917 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -9.2337 Å / Origin y: -5.2763 Å / Origin z: 13.4028 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller











 PDBj
PDBj





