[English] 日本語
 Yorodumi
Yorodumi- PDB-6eem: Crystal structure of Papaver somniferum tyrosine decarboxylase in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6eem | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | ||||||
 Components Components | (Tyrosine/dopa ...) x 2 | ||||||
 Keywords Keywords |  LYASE / LYASE /  Aromatic Amino Acid Decarboxylase Aromatic Amino Acid Decarboxylase | ||||||
| Function / homology |  Function and homology information Function and homology information tyrosine decarboxylase / tyrosine decarboxylase /  tyrosine decarboxylase activity / amino acid metabolic process / tyrosine decarboxylase activity / amino acid metabolic process /  pyridoxal phosphate binding pyridoxal phosphate bindingSimilarity search - Function | ||||||
| Biological species |   Papaver somniferum (opium poppy) Papaver somniferum (opium poppy) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.61000662305 Å MOLECULAR REPLACEMENT / Resolution: 2.61000662305 Å | ||||||
 Authors Authors | Torrens-Spence, M.P. / Chiang, Y. / Smith, T. / Vicent, M.A. / Wang, Y. / Weng, J.K. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2020 Journal: Proc.Natl.Acad.Sci.USA / Year: 2020Title: Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins. Authors: Torrens-Spence, M.P. / Chiang, Y.C. / Smith, T. / Vicent, M.A. / Wang, Y. / Weng, J.K. #1:  Journal: Biorxiv / Year: 2019 Journal: Biorxiv / Year: 2019Title: Structural basis for independent origins of new catalytic machineries in plant AAAD proteins Authors: Torrens-Spence, M.P. / Chiang, Y.-C. / Smith, T. / Vicent, M.A. / Wang, Y. / Weng, J.K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6eem.cif.gz 6eem.cif.gz | 440.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6eem.ent.gz pdb6eem.ent.gz | 323.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6eem.json.gz 6eem.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ee/6eem https://data.pdbj.org/pub/pdb/validation_reports/ee/6eem ftp://data.pdbj.org/pub/pdb/validation_reports/ee/6eem ftp://data.pdbj.org/pub/pdb/validation_reports/ee/6eem | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6eeiC  6eeqC  6eewC  1js3S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Tyrosine/dopa ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 56613.719 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Papaver somniferum (opium poppy) / Gene: tydc9 / Production host: Papaver somniferum (opium poppy) / Gene: tydc9 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: O82415,  aromatic-L-amino-acid decarboxylase aromatic-L-amino-acid decarboxylase |
|---|---|
| #2: Protein | Mass: 56841.828 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Papaver somniferum (opium poppy) / Gene: tydc9 / Production host: Papaver somniferum (opium poppy) / Gene: tydc9 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: O82415,  aromatic-L-amino-acid decarboxylase aromatic-L-amino-acid decarboxylase |
-Non-polymers , 4 types, 261 molecules 

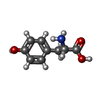




| #3: Chemical | ChemComp-0PR / | ||||
|---|---|---|---|---|---|
| #4: Chemical | ChemComp-SO4 /  Sulfate Sulfate#5: Chemical | ChemComp-TYR / |  Tyrosine Tyrosine#6: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.9 Å3/Da / Density % sol: 57.58 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 5 Details: 1.2 M ammonium Sulfate 0.1 Bis Tris pH 5.0 and 1% w/v PEG 3350 |
-Data collection
| Diffraction | Mean temperature: 80 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-E / Wavelength: 0.97918 Å / Beamline: 24-ID-E / Wavelength: 0.97918 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Nov 5, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97918 Å / Relative weight: 1 : 0.97918 Å / Relative weight: 1 |
| Reflection | Resolution: 2.61→167 Å / Num. obs: 39550 / % possible obs: 100 % / Redundancy: 8.4 % / Biso Wilson estimate: 34.2140174036 Å2 / CC1/2: 0.985 / Net I/σ(I): 9.1 |
| Reflection shell | Resolution: 2.61→2.75 Å / Redundancy: 8.5 % / Mean I/σ(I) obs: 1.6 / Num. unique obs: 5654 / CC1/2: 0.567 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1JS3 Resolution: 2.61000662305→77.026298568 Å / SU ML: 0.337130549857 / Cross valid method: THROUGHOUT / σ(F): 1.32602802391 / Phase error: 23.3974781119
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 37.7978677589 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.61000662305→77.026298568 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj




