[English] 日本語
 Yorodumi
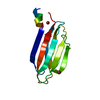
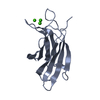
Yorodumi- PDB-6cxw: Synaptotagmin 1 C2A beta4 chimera with enhanced PIP2 binding function -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6cxw | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Synaptotagmin 1 C2A beta4 chimera with enhanced PIP2 binding function | |||||||||
 Components Components | Synaptotagmin 1 C2A beta4 chimera | |||||||||
 Keywords Keywords |  EXOCYTOSIS / EXOCYTOSIS /  C2 domain C2 domain | |||||||||
| Function / homology |  Function and homology information Function and homology informationsynchronous neurotransmitter secretion / fast, calcium ion-dependent exocytosis of neurotransmitter / positive regulation of calcium ion-dependent exocytosis of neurotransmitter /  syntaxin-3 binding / calcium-dependent activation of synaptic vesicle fusion / regulation of regulated secretory pathway / calcium ion sensor activity / spontaneous neurotransmitter secretion / Glutamate Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle ...synchronous neurotransmitter secretion / fast, calcium ion-dependent exocytosis of neurotransmitter / positive regulation of calcium ion-dependent exocytosis of neurotransmitter / syntaxin-3 binding / calcium-dependent activation of synaptic vesicle fusion / regulation of regulated secretory pathway / calcium ion sensor activity / spontaneous neurotransmitter secretion / Glutamate Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle ...synchronous neurotransmitter secretion / fast, calcium ion-dependent exocytosis of neurotransmitter / positive regulation of calcium ion-dependent exocytosis of neurotransmitter /  syntaxin-3 binding / calcium-dependent activation of synaptic vesicle fusion / regulation of regulated secretory pathway / calcium ion sensor activity / spontaneous neurotransmitter secretion / Glutamate Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle / Acetylcholine Neurotransmitter Release Cycle / Serotonin Neurotransmitter Release Cycle / dense core granule / GABA synthesis, release, reuptake and degradation / positive regulation of vesicle fusion / chromaffin granule membrane / Dopamine Neurotransmitter Release Cycle / calcium ion-regulated exocytosis of neurotransmitter / positive regulation of calcium ion-dependent exocytosis / vesicle docking / regulation of calcium ion-dependent exocytosis / exocytic vesicle / positive regulation of dopamine secretion / protein heterooligomerization / Cargo recognition for clathrin-mediated endocytosis / syntaxin-3 binding / calcium-dependent activation of synaptic vesicle fusion / regulation of regulated secretory pathway / calcium ion sensor activity / spontaneous neurotransmitter secretion / Glutamate Neurotransmitter Release Cycle / Norepinephrine Neurotransmitter Release Cycle / Acetylcholine Neurotransmitter Release Cycle / Serotonin Neurotransmitter Release Cycle / dense core granule / GABA synthesis, release, reuptake and degradation / positive regulation of vesicle fusion / chromaffin granule membrane / Dopamine Neurotransmitter Release Cycle / calcium ion-regulated exocytosis of neurotransmitter / positive regulation of calcium ion-dependent exocytosis / vesicle docking / regulation of calcium ion-dependent exocytosis / exocytic vesicle / positive regulation of dopamine secretion / protein heterooligomerization / Cargo recognition for clathrin-mediated endocytosis /  Clathrin-mediated endocytosis / calcium-ion regulated exocytosis / neurotransmitter secretion / positive regulation of dendrite extension / calcium-dependent phospholipid binding / neuron projection terminus / Clathrin-mediated endocytosis / calcium-ion regulated exocytosis / neurotransmitter secretion / positive regulation of dendrite extension / calcium-dependent phospholipid binding / neuron projection terminus /  syntaxin-1 binding / syntaxin-1 binding /  syntaxin binding / regulation of synaptic vesicle exocytosis / low-density lipoprotein particle receptor binding / syntaxin binding / regulation of synaptic vesicle exocytosis / low-density lipoprotein particle receptor binding /  clathrin binding / clathrin binding /  phosphatidylserine binding / regulation of dopamine secretion / synaptic vesicle exocytosis / phosphatidylserine binding / regulation of dopamine secretion / synaptic vesicle exocytosis /  excitatory synapse / synaptic vesicle endocytosis / detection of calcium ion / positive regulation of synaptic transmission / excitatory synapse / synaptic vesicle endocytosis / detection of calcium ion / positive regulation of synaptic transmission /  regulation of synaptic transmission, glutamatergic / regulation of synaptic transmission, glutamatergic /  phosphatidylinositol-4,5-bisphosphate binding / hippocampal mossy fiber to CA3 synapse / cellular response to calcium ion / phosphatidylinositol-4,5-bisphosphate binding / hippocampal mossy fiber to CA3 synapse / cellular response to calcium ion /  SNARE binding / SNARE binding /  secretory granule / secretory granule /  phospholipid binding / phospholipid binding /  terminal bouton / synaptic vesicle membrane / response to calcium ion / calcium-dependent protein binding / terminal bouton / synaptic vesicle membrane / response to calcium ion / calcium-dependent protein binding /  synaptic vesicle / synaptic vesicle /  presynaptic membrane / presynaptic membrane /  cell differentiation / cell differentiation /  calmodulin binding / neuron projection / protein heterodimerization activity / calmodulin binding / neuron projection / protein heterodimerization activity /  axon / glutamatergic synapse / axon / glutamatergic synapse /  calcium ion binding / calcium ion binding /  Golgi apparatus / identical protein binding / Golgi apparatus / identical protein binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.832 Å MOLECULAR REPLACEMENT / Resolution: 1.832 Å | |||||||||
 Authors Authors | Sutton, R.B. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Engineering a C2-domain to bind PIP2 Authors: Evans, C.S. / Lou, X. / Bai, H. / Rice, A.M. / Salcido-Holguin, L. / Rodriguez, E.N. / Sutton, R.B. / Chapman, E.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6cxw.cif.gz 6cxw.cif.gz | 91.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6cxw.ent.gz pdb6cxw.ent.gz | 70.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6cxw.json.gz 6cxw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cx/6cxw https://data.pdbj.org/pub/pdb/validation_reports/cx/6cxw ftp://data.pdbj.org/pub/pdb/validation_reports/cx/6cxw ftp://data.pdbj.org/pub/pdb/validation_reports/cx/6cxw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4weeS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14692.845 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: Syt1 / Production host: Rattus norvegicus (Norway rat) / Gene: Syt1 / Production host:   Escherichia coli (E. coli) / References: UniProt: P21707 Escherichia coli (E. coli) / References: UniProt: P21707 |
|---|---|
| #2: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.86 Å3/Da / Density % sol: 33.86 % |
|---|---|
Crystal grow | Temperature: 300 K / Method: vapor diffusion / pH: 8.5 / Details: 50% PEG-4000, 0.4 mM Sodium acetate, 100 mM Tris |
-Data collection
| Diffraction | Mean temperature: 90 K | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL7-1 / Wavelength: 1.283572 Å / Beamline: BL7-1 / Wavelength: 1.283572 Å | ||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jan 12, 2016 / Details: Rh coasted flat bent mirror | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1.283572 Å / Relative weight: 1 : 1.283572 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.83→31.73 Å / Num. obs: 9789 / % possible obs: 96.8 % / Redundancy: 8.7 % / CC1/2: 0.999 / Rmerge(I) obs: 0.034 / Rpim(I) all: 0.011 / Rrim(I) all: 0.036 / Net I/σ(I): 51.6 / Num. measured all: 85169 / Scaling rejects: 88 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4wee Resolution: 1.832→31.728 Å / SU ML: 0.15 / Cross valid method: THROUGHOUT / σ(F): 1.39 / Phase error: 19.59 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 110.79 Å2 / Biso mean: 24.2284 Å2 / Biso min: 5.97 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.832→31.728 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 7
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj





