[English] 日本語
 Yorodumi
Yorodumi- PDB-5tjz: Structure of 4-Hydroxytetrahydrodipicolinate Reductase from Mycob... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5tjz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of 4-Hydroxytetrahydrodipicolinate Reductase from Mycobacterium tuberculosis with NADPH and 2,6 Pyridine Dicarboxylic Acid | ||||||
 Components Components | 4-hydroxy-tetrahydrodipicolinate reductase | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  Lysine Biosynthesis Lysine Biosynthesis | ||||||
| Function / homology |  Function and homology information Function and homology information 4-hydroxy-tetrahydrodipicolinate reductase / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor / 4-hydroxy-tetrahydrodipicolinate reductase / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor /  4-hydroxy-tetrahydrodipicolinate reductase / 4-hydroxy-tetrahydrodipicolinate reductase /  NADH binding / diaminopimelate biosynthetic process / lysine biosynthetic process via diaminopimelate / NADH binding / diaminopimelate biosynthetic process / lysine biosynthetic process via diaminopimelate /  NADPH binding / peptidoglycan-based cell wall / NAD binding / NADPH binding / peptidoglycan-based cell wall / NAD binding /  NADP binding ... NADP binding ... 4-hydroxy-tetrahydrodipicolinate reductase / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor / 4-hydroxy-tetrahydrodipicolinate reductase / oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor /  4-hydroxy-tetrahydrodipicolinate reductase / 4-hydroxy-tetrahydrodipicolinate reductase /  NADH binding / diaminopimelate biosynthetic process / lysine biosynthetic process via diaminopimelate / NADH binding / diaminopimelate biosynthetic process / lysine biosynthetic process via diaminopimelate /  NADPH binding / peptidoglycan-based cell wall / NAD binding / NADPH binding / peptidoglycan-based cell wall / NAD binding /  NADP binding / NADP binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Mycobacterium tuberculosis (bacteria) Mycobacterium tuberculosis (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Mank, N.J. / Arnette, A.K. / Klapper, V. / Chruszcz, M. | ||||||
 Citation Citation |  Journal: Biochim Biophys Acta Gen Subj / Year: 2021 Journal: Biochim Biophys Acta Gen Subj / Year: 2021Title: Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus. Authors: Pote, S. / Kachhap, S. / Mank, N.J. / Daneshian, L. / Klapper, V. / Pye, S. / Arnette, A.K. / Shimizu, L.S. / Borowski, T. / Chruszcz, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5tjz.cif.gz 5tjz.cif.gz | 121.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5tjz.ent.gz pdb5tjz.ent.gz | 91.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5tjz.json.gz 5tjz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tj/5tjz https://data.pdbj.org/pub/pdb/validation_reports/tj/5tjz ftp://data.pdbj.org/pub/pdb/validation_reports/tj/5tjz ftp://data.pdbj.org/pub/pdb/validation_reports/tj/5tjz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5tejC  5tekC  5temC  5tenC  5tjyC  5ugvC  5us6C  1c3vS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein |  / HTPA reductase / HTPA reductaseMass: 25682.262 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mycobacterium tuberculosis (strain ATCC 25177 / H37Ra) (bacteria) Mycobacterium tuberculosis (strain ATCC 25177 / H37Ra) (bacteria)Strain: ATCC 25177 / H37Ra / Gene: dapB, MRA_2798 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) Escherichia coli (E. coli) / Strain (production host): BL21(DE3)References: UniProt: A5U6C6, UniProt: P9WP23*PLUS,  4-hydroxy-tetrahydrodipicolinate reductase 4-hydroxy-tetrahydrodipicolinate reductase |
|---|
-Non-polymers , 9 types, 332 molecules 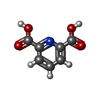















| #2: Chemical | ChemComp-PDC /  Dipicolinic acid Dipicolinic acid | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-BEZ /  Benzoic acid Benzoic acid | ||||||
| #4: Chemical | ChemComp-IMD /  Imidazole Imidazole | ||||||
| #5: Chemical | ChemComp-NAP /  Nicotinamide adenine dinucleotide phosphate Nicotinamide adenine dinucleotide phosphate | ||||||
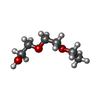
| #6: Chemical | ChemComp-AE3 /  2-(2-Ethoxyethoxy)ethanol 2-(2-Ethoxyethoxy)ethanol | ||||||
| #7: Chemical |  Chloride Chloride#8: Chemical | ChemComp-EDO /  Ethylene glycol Ethylene glycol#9: Chemical | ChemComp-SO4 / |  Sulfate Sulfate#10: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.19 Å3/Da / Density % sol: 61.44 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop Details: 0.1 M Sodium Chloride, 0.1 M Bis-Tris pH 6.5, 1.5 M Ammonium Sulfate |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: Cryostream | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 1 Å / Beamline: 19-ID / Wavelength: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Aug 8, 2014 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.5→30 Å / Num. obs: 52243 / % possible obs: 95.1 % / Redundancy: 7.6 % / Rmerge(I) obs: 0.054 / Net I/av σ(I): 42.436 / Net I/σ(I): 11.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1C3V Resolution: 1.5→30 Å / Cor.coef. Fo:Fc: 0.976 / Cor.coef. Fo:Fc free: 0.97 / SU B: 1.744 / SU ML: 0.035 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.055 / ESU R Free: 0.057 Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT U VALUES : WITH TLS ADDED
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 92.19 Å2 / Biso mean: 29.958 Å2 / Biso min: 17.13 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.5→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.501→1.54 Å / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj












