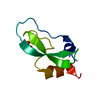
Entry Database : PDB / ID : 5o94Title X-ray structure of a zinc binding GB1 mutant Metal binding GB1 Keywords / Function / homology / / / Biological species Streptococcus sp. (bacteria)Method / / / Resolution : 1.9 Å Authors Rothlisberger, U. / Bozkurt, E. / Hovius, R. / Perez, M.A.S. / Browning, N.J. Funding support Organization Grant number Country Swiss National Science Foundation 200020-146645
Journal : J. Am. Chem. Soc. / Year : 2018Title : Genetic Algorithm Based Design and Experimental Characterization of a Highly Thermostable Metalloprotein.Authors : Bozkurt, E. / Perez, M.A.S. / Hovius, R. / Browning, N.J. / Rothlisberger, U. History Deposition Jun 15, 2017 Deposition site / Processing site Revision 1.0 Apr 4, 2018 Provider / Type Revision 1.1 Apr 11, 2018 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Structure summary Category chem_comp / citation ... chem_comp / citation / entity / pdbx_entity_nonpoly Item _chem_comp.name / _citation.journal_volume ... _chem_comp.name / _citation.journal_volume / _citation.page_first / _citation.page_last / _entity.pdbx_description / _pdbx_entity_nonpoly.name Revision 1.2 Jan 17, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / entity / pdbx_entity_nonpoly / pdbx_initial_refinement_model / struct_conn Item _chem_comp.name / _database_2.pdbx_DOI ... _chem_comp.name / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _entity.pdbx_description / _pdbx_entity_nonpoly.name / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_symmetry
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Streptococcus sp. (bacteria)
Streptococcus sp. (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å
MOLECULAR REPLACEMENT / Resolution: 1.9 Å  Authors
Authors Switzerland, 1items
Switzerland, 1items  Citation
Citation Journal: J. Am. Chem. Soc. / Year: 2018
Journal: J. Am. Chem. Soc. / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5o94.cif.gz
5o94.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5o94.ent.gz
pdb5o94.ent.gz PDB format
PDB format 5o94.json.gz
5o94.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/o9/5o94
https://data.pdbj.org/pub/pdb/validation_reports/o9/5o94 ftp://data.pdbj.org/pub/pdb/validation_reports/o9/5o94
ftp://data.pdbj.org/pub/pdb/validation_reports/o9/5o94

 Links
Links Assembly
Assembly
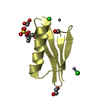
 Components
Components Streptococcus sp. (bacteria) / Production host:
Streptococcus sp. (bacteria) / Production host: 
 Escherichia coli BL21(DE3) (bacteria)
Escherichia coli BL21(DE3) (bacteria)












 2-Methyl-2,4-pentanediol
2-Methyl-2,4-pentanediol Sulfate
Sulfate Ethylene glycol
Ethylene glycol Glycerol
Glycerol Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X06DA / Wavelength: 1 Å
/ Beamline: X06DA / Wavelength: 1 Å : 1 Å / Relative weight: 1
: 1 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj




