| Deposited unit | C: Integrin alpha-2
A: Integrin alpha-2
B: Integrin alpha-2
D: Integrin alpha-2
E: Integrin alpha-2
F: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 130,805 | 21 |
|---|
| Polymers | 129,885 | 6 |
|---|
| Non-polymers | 921 | 15 |
|---|
| Water | 6,557 | 364 |
|---|
|
|---|
| 1 | C: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 21,688 | 2 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 40 | 1 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| 2 | A: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 21,961 | 6 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 313 | 5 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| 3 | B: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 21,723 | 3 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 76 | 2 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| 4 | D: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 22,007 | 6 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 360 | 5 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| 5 | E: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 21,688 | 2 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 40 | 1 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| 6 | F: Integrin alpha-2
hetero molecules
| Theoretical mass | Number of molelcules |
|---|
| Total (without water) | 21,740 | 2 |
|---|
| Polymers | 21,647 | 1 |
|---|
| Non-polymers | 92 | 1 |
|---|
| Water | 18 | 1 |
|---|
| Type | Name | Symmetry operation | Number |
|---|
| identity operation | 1_555 | x,y,z | 1 |
|
|---|
| Unit cell | | Length a, b, c (Å) | 144.066, 144.066, 130.004 |
|---|
| Angle α, β, γ (deg.) | 90.00, 90.00, 90.00 |
|---|
| Int Tables number | 96 |
|---|
| Space group name H-M | P43212 |
|---|
|
|---|
| Components on special symmetry positions | | ID | Model | Components |
|---|
| 1 | 1 | E-159-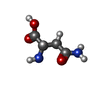 ASN | | 2 | 1 | A-302- CL | | 3 | 1 | F-301- GOL | | 4 | 1 | F-441- HOH | | 5 | 1 | F-443- HOH |
|
|---|
| Noncrystallographic symmetry (NCS) | NCS domain: | ID | Ens-ID | Details |
|---|
| 1 | 1 | (chain A and (resseq 7:20 or (resid 21 and (name...| 2 | 1 | (chain B and (resseq 7:20 or (resid 21 and (name...| 3 | 1 | (chain C and (resseq 7:20 or (resid 21 and (name...| 4 | 1 | (chain D and (resseq 7:20 or (resid 21 and (name...| 5 | 1 | (chain E and (resseq 7:20 or (resid 21 and (name...| 6 | 1 | (chain F and (resseq 7:20 or (resid 21 and (name... | | | | | |
NCS domain segments: Ens-ID: 1 | Dom-ID | Component-ID | Beg auth comp-ID | Beg label comp-ID | End auth comp-ID | End label comp-ID | Selection details | Auth asym-ID | Label asym-ID | Auth seq-ID | Label seq-ID |
|---|
| 1 | 1 | LEULEUILEILE(chain A and (resseq 7:20 or (resid 21 and (name...AB| 7 - 20 | 3 - 16 | | 1 | 2 | TYRTYRTYRTYR(chain A and (resseq 7:20 or (resid 21 and (name...AB| 21 | 17 | | 1 | 3 | SERSERGLYGLY(chain A and (resseq 7:20 or (resid 21 and (name...AB| 6 - 201 | 2 - 197 | | 1 | 4 | SERSERGLYGLY(chain A and (resseq 7:20 or (resid 21 and (name...AB| 6 - 201 | 2 - 197 | | 1 | 5 | SERSERGLYGLY(chain A and (resseq 7:20 or (resid 21 and (name...AB| 6 - 201 | 2 - 197 | | 1 | 6 | SERSERGLYGLY| (chain A and (resseq 7:20 or (resid 21 and (name... | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords CELL ADHESION /
CELL ADHESION /  Adhesion /
Adhesion /  signaling
signaling Function and homology information
Function and homology information collagen receptor activity / substrate-dependent cell migration / positive regulation of transmission of nerve impulse / positive regulation of cell projection organization / collagen binding involved in cell-matrix adhesion / integrin alpha2-beta1 complex / response to parathyroid hormone / hypotonic response / response to L-ascorbic acid / skin morphogenesis ...
collagen receptor activity / substrate-dependent cell migration / positive regulation of transmission of nerve impulse / positive regulation of cell projection organization / collagen binding involved in cell-matrix adhesion / integrin alpha2-beta1 complex / response to parathyroid hormone / hypotonic response / response to L-ascorbic acid / skin morphogenesis ... collagen receptor activity / substrate-dependent cell migration / positive regulation of transmission of nerve impulse / positive regulation of cell projection organization / collagen binding involved in cell-matrix adhesion / integrin alpha2-beta1 complex / response to parathyroid hormone / hypotonic response / response to L-ascorbic acid / skin morphogenesis / CHL1 interactions / Laminin interactions / basal part of cell / positive regulation of smooth muscle contraction / positive regulation of phagocytosis, engulfment / collagen-activated signaling pathway / Platelet Adhesion to exposed collagen /
collagen receptor activity / substrate-dependent cell migration / positive regulation of transmission of nerve impulse / positive regulation of cell projection organization / collagen binding involved in cell-matrix adhesion / integrin alpha2-beta1 complex / response to parathyroid hormone / hypotonic response / response to L-ascorbic acid / skin morphogenesis / CHL1 interactions / Laminin interactions / basal part of cell / positive regulation of smooth muscle contraction / positive regulation of phagocytosis, engulfment / collagen-activated signaling pathway / Platelet Adhesion to exposed collagen /  mammary gland development / hepatocyte differentiation / mesodermal cell differentiation /
mammary gland development / hepatocyte differentiation / mesodermal cell differentiation /  focal adhesion assembly /
focal adhesion assembly /  heparan sulfate proteoglycan binding / response to muscle activity /
heparan sulfate proteoglycan binding / response to muscle activity /  integrin complex / positive regulation of positive chemotaxis / positive regulation of leukocyte migration / cell adhesion mediated by integrin / MET activates PTK2 signaling / Syndecan interactions / positive regulation of epithelial cell migration / cell-substrate adhesion / positive regulation of smooth muscle cell migration / response to amine / positive regulation of cell adhesion / ECM proteoglycans / positive regulation of collagen biosynthetic process / Integrin cell surface interactions /
integrin complex / positive regulation of positive chemotaxis / positive regulation of leukocyte migration / cell adhesion mediated by integrin / MET activates PTK2 signaling / Syndecan interactions / positive regulation of epithelial cell migration / cell-substrate adhesion / positive regulation of smooth muscle cell migration / response to amine / positive regulation of cell adhesion / ECM proteoglycans / positive regulation of collagen biosynthetic process / Integrin cell surface interactions /  laminin binding / detection of mechanical stimulus involved in sensory perception of pain / axon terminus /
laminin binding / detection of mechanical stimulus involved in sensory perception of pain / axon terminus /  collagen binding / cell-matrix adhesion / positive regulation of translation / female pregnancy / cellular response to estradiol stimulus / integrin-mediated signaling pathway / animal organ morphogenesis / positive regulation of smooth muscle cell proliferation /
collagen binding / cell-matrix adhesion / positive regulation of translation / female pregnancy / cellular response to estradiol stimulus / integrin-mediated signaling pathway / animal organ morphogenesis / positive regulation of smooth muscle cell proliferation /  cell-cell adhesion / cellular response to mechanical stimulus /
cell-cell adhesion / cellular response to mechanical stimulus /  blood coagulation /
blood coagulation /  integrin binding / virus receptor activity /
integrin binding / virus receptor activity /  amyloid-beta binding / response to hypoxia /
amyloid-beta binding / response to hypoxia /  cell adhesion / response to xenobiotic stimulus / external side of plasma membrane /
cell adhesion / response to xenobiotic stimulus / external side of plasma membrane /  focal adhesion / protein-containing complex binding / perinuclear region of cytoplasm /
focal adhesion / protein-containing complex binding / perinuclear region of cytoplasm /  cell surface /
cell surface /  metal ion binding /
metal ion binding /  plasma membrane
plasma membrane
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.153 Å
SYNCHROTRON / Resolution: 2.153 Å  Authors
Authors Citation
Citation Journal: Sci Rep / Year: 2018
Journal: Sci Rep / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5hj2.cif.gz
5hj2.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5hj2.ent.gz
pdb5hj2.ent.gz PDB format
PDB format 5hj2.json.gz
5hj2.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/hj/5hj2
https://data.pdbj.org/pub/pdb/validation_reports/hj/5hj2 ftp://data.pdbj.org/pub/pdb/validation_reports/hj/5hj2
ftp://data.pdbj.org/pub/pdb/validation_reports/hj/5hj2 Links
Links Assembly
Assembly






 Movie
Movie Controller
Controller













 PDBj
PDBj












