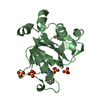
Entry Database : PDB / ID : 3gxbTitle Crystal structure of VWF A2 domain von Willebrand factor Keywords / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / / Resolution : 1.9 Å Authors Zhou, Y.F. / Springer, T.A. Journal : Proc.Natl.Acad.Sci.USA / Year : 2009Title : Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.Authors : Zhang, Q. / Zhou, Y.F. / Zhang, C.Z. / Zhang, X. / Lu, C. / Springer, T.A. History Deposition Apr 2, 2009 Deposition site / Processing site Revision 1.0 May 5, 2009 Provider / Type Revision 1.1 Jul 13, 2011 Group / Version format complianceRevision 2.0 Jul 29, 2020 Group Atomic model / Data collection ... Atomic model / Data collection / Database references / Derived calculations / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / entity / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / struct_asym / struct_conn / struct_ref_seq_dif / struct_site / struct_site_gen Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_seq_id / _atom_site.label_asym_id / _atom_site.label_entity_id / _atom_site.occupancy / _chem_comp.name / _chem_comp.type / _pdbx_entity_nonpoly.entity_id / _pdbx_entity_nonpoly.name / _pdbx_struct_assembly_gen.asym_id_list / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_role / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_ref_seq_dif.details Description / Provider / Type Revision 2.1 Sep 6, 2023 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim Item _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ... _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components
 Keywords
Keywords CELL ADHESION / VWA-like fold /
CELL ADHESION / VWA-like fold /  Blood coagulation / Cleavage on pair of basic residues / Disease mutation /
Blood coagulation / Cleavage on pair of basic residues / Disease mutation /  Disulfide bond /
Disulfide bond /  Extracellular matrix /
Extracellular matrix /  Glycoprotein /
Glycoprotein /  Hemostasis /
Hemostasis /  Isopeptide bond /
Isopeptide bond /  Secreted /
Secreted /  von Willebrand disease
von Willebrand disease Function and homology information
Function and homology information Weibel-Palade body / Defective F8 binding to von Willebrand factor / Enhanced binding of GP1BA variant to VWF multimer:collagen / Defective binding of VWF variant to GPIb:IX:V /
Weibel-Palade body / Defective F8 binding to von Willebrand factor / Enhanced binding of GP1BA variant to VWF multimer:collagen / Defective binding of VWF variant to GPIb:IX:V /  hemostasis /
hemostasis /  platelet alpha granule / Platelet Adhesion to exposed collagen ...Enhanced cleavage of VWF variant by ADAMTS13 / Defective VWF cleavage by ADAMTS13 variant / Defective VWF binding to collagen type I /
platelet alpha granule / Platelet Adhesion to exposed collagen ...Enhanced cleavage of VWF variant by ADAMTS13 / Defective VWF cleavage by ADAMTS13 variant / Defective VWF binding to collagen type I /  Weibel-Palade body / Defective F8 binding to von Willebrand factor / Enhanced binding of GP1BA variant to VWF multimer:collagen / Defective binding of VWF variant to GPIb:IX:V /
Weibel-Palade body / Defective F8 binding to von Willebrand factor / Enhanced binding of GP1BA variant to VWF multimer:collagen / Defective binding of VWF variant to GPIb:IX:V /  hemostasis /
hemostasis /  platelet alpha granule / Platelet Adhesion to exposed collagen / positive regulation of intracellular signal transduction / GP1b-IX-V activation signalling / p130Cas linkage to MAPK signaling for integrins / cell-substrate adhesion / Defective F8 cleavage by thrombin / Platelet Aggregation (Plug Formation) /
platelet alpha granule / Platelet Adhesion to exposed collagen / positive regulation of intracellular signal transduction / GP1b-IX-V activation signalling / p130Cas linkage to MAPK signaling for integrins / cell-substrate adhesion / Defective F8 cleavage by thrombin / Platelet Aggregation (Plug Formation) /  immunoglobulin binding / GRB2:SOS provides linkage to MAPK signaling for Integrins / Integrin cell surface interactions /
immunoglobulin binding / GRB2:SOS provides linkage to MAPK signaling for Integrins / Integrin cell surface interactions /  collagen binding / Intrinsic Pathway of Fibrin Clot Formation / Integrin signaling /
collagen binding / Intrinsic Pathway of Fibrin Clot Formation / Integrin signaling /  extracellular matrix / platelet alpha granule lumen / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation /
extracellular matrix / platelet alpha granule lumen / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation /  platelet activation / response to wounding / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / Signaling by BRAF and RAF1 fusions /
platelet activation / response to wounding / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / Signaling by BRAF and RAF1 fusions /  blood coagulation /
blood coagulation /  integrin binding / Platelet degranulation / protein-folding chaperone binding / collagen-containing extracellular matrix /
integrin binding / Platelet degranulation / protein-folding chaperone binding / collagen-containing extracellular matrix /  protease binding /
protease binding /  cell adhesion /
cell adhesion /  endoplasmic reticulum /
endoplasmic reticulum /  extracellular space / extracellular exosome / extracellular region / identical protein binding
extracellular space / extracellular exosome / extracellular region / identical protein binding
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.9 Å
molecular replacement / Resolution: 1.9 Å  Authors
Authors Citation
Citation Journal: Proc.Natl.Acad.Sci.USA / Year: 2009
Journal: Proc.Natl.Acad.Sci.USA / Year: 2009 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3gxb.cif.gz
3gxb.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3gxb.ent.gz
pdb3gxb.ent.gz PDB format
PDB format 3gxb.json.gz
3gxb.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/gx/3gxb
https://data.pdbj.org/pub/pdb/validation_reports/gx/3gxb ftp://data.pdbj.org/pub/pdb/validation_reports/gx/3gxb
ftp://data.pdbj.org/pub/pdb/validation_reports/gx/3gxb
 Links
Links Assembly
Assembly


 Components
Components / vWF / von Willebrand antigen 2 / von Willebrand antigen II
/ vWF / von Willebrand antigen 2 / von Willebrand antigen II
 Homo sapiens (human) / Gene: F8VWF, VWF / Plasmid: pLEXm / References: UniProt: P04275
Homo sapiens (human) / Gene: F8VWF, VWF / Plasmid: pLEXm / References: UniProt: P04275 / Mass: 424.401 Da / Num. of mol.: 2
/ Mass: 424.401 Da / Num. of mol.: 2 N-Acetylglucosamine
N-Acetylglucosamine Sulfate
Sulfate Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 24-ID-E / Wavelength: 0.9795
/ Beamline: 24-ID-E / Wavelength: 0.9795  : 0.9795 Å / Relative weight: 1
: 0.9795 Å / Relative weight: 1 
 molecular replacement
molecular replacement Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller












 PDBj
PDBj


















