[English] 日本語
 Yorodumi
Yorodumi- PDB-5dwk: Diacylglycerol Kinase solved by multi crystal multi orientation n... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5dwk | ||||||
|---|---|---|---|---|---|---|---|
| Title | Diacylglycerol Kinase solved by multi crystal multi orientation native SAD | ||||||
 Components Components | Diacylglycerol kinase | ||||||
 Keywords Keywords |  TRANSFERASE / native SAD / DAGK / multi crystal / multi orientation TRANSFERASE / native SAD / DAGK / multi crystal / multi orientation | ||||||
| Function / homology |  Function and homology information Function and homology informationdiacylglycerol kinase (ATP) / ATP-dependent diacylglycerol kinase activity / phosphatidic acid biosynthetic process / response to UV /  phosphorylation / phosphorylation /  ATP binding / ATP binding /  membrane / identical protein binding / membrane / identical protein binding /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.601 Å SAD / Resolution: 2.601 Å | ||||||
 Authors Authors | Weinert, T. / Olieric, V. / Finke, A.D. / Li, D. / Caffrey, M. / Wang, M. | ||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2016 Journal: Acta Crystallogr D Struct Biol / Year: 2016Title: Data-collection strategy for challenging native SAD phasing. Authors: Olieric, V. / Weinert, T. / Finke, A.D. / Anders, C. / Li, D. / Olieric, N. / Borca, C.N. / Steinmetz, M.O. / Caffrey, M. / Jinek, M. / Wang, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5dwk.cif.gz 5dwk.cif.gz | 238.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5dwk.ent.gz pdb5dwk.ent.gz | 195 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5dwk.json.gz 5dwk.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dw/5dwk https://data.pdbj.org/pub/pdb/validation_reports/dw/5dwk ftp://data.pdbj.org/pub/pdb/validation_reports/dw/5dwk ftp://data.pdbj.org/pub/pdb/validation_reports/dw/5dwk | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 6 molecules ABCDEF
| #1: Protein |  / DAGK / Diglyceride kinase / DGK / DAGK / Diglyceride kinase / DGKMass: 14204.451 Da / Num. of mol.: 6 / Fragment: UNP residues 2-122 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli (E. coli) / Strain: K12 / Gene: dgkA, b4042, JW4002 / Plasmid: PTRCHISB_DGKA_DELTA7 / Production host: Escherichia coli (E. coli) / Strain: K12 / Gene: dgkA, b4042, JW4002 / Plasmid: PTRCHISB_DGKA_DELTA7 / Production host:   Escherichia coli (E. coli) / Strain (production host): Wh1061 / References: UniProt: P0ABN1, diacylglycerol kinase (ATP) Escherichia coli (E. coli) / Strain (production host): Wh1061 / References: UniProt: P0ABN1, diacylglycerol kinase (ATP) |
|---|
-Non-polymers , 7 types, 53 molecules 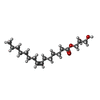
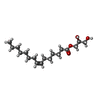











| #2: Chemical | ChemComp-78N / ( #3: Chemical | ChemComp-78M / ( #4: Chemical | ChemComp-NA / | #5: Chemical | ChemComp-ZN / | #6: Chemical | ChemComp-CIT / |  Citric acid Citric acid#7: Chemical | ChemComp-ACT / |  Acetate Acetate#8: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.91 Å3/Da / Density % sol: 57.68 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: lipidic cubic phase / pH: 5.6 Details: 3-5 %(V/V) 2-METHYL-2, 4-PENTANEDIOL, 0.1 M SODIUM CHLORIDE, 0.06 M MAGNESIUM ACETATE, 0.1 M SODIUM CITRATE/HCL PH 5.6. CRYSTALLIZED USING THE IN MESO (LIPIDIC CUBIC PHASE) METHOD AT 4 ...Details: 3-5 %(V/V) 2-METHYL-2, 4-PENTANEDIOL, 0.1 M SODIUM CHLORIDE, 0.06 M MAGNESIUM ACETATE, 0.1 M SODIUM CITRATE/HCL PH 5.6. CRYSTALLIZED USING THE IN MESO (LIPIDIC CUBIC PHASE) METHOD AT 4 DEGREE CELSIUS WITH THE 7.8 MONOACYLGLYCEROL (7.8 MAG) AS THE HOSTING LIPID. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 2.0664 Å / Beamline: X06DA / Wavelength: 2.0664 Å |
| Detector | Type: DECTRIS PILATUS 2M-F / Detector: PIXEL / Date: Jun 14, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 2.0664 Å / Relative weight: 1 : 2.0664 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→50 Å / Num. obs: 58770 / % possible obs: 99.6 % / Redundancy: 142.9 % / Rmerge(I) obs: 0.221 / Net I/σ(I): 36.46 |
| Reflection shell | Resolution: 2.6→2.67 Å / Redundancy: 55.85 % / Rmerge(I) obs: 1.13 / Mean I/σ(I) obs: 4.18 / % possible all: 96.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 2.601→45.785 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 1.93 / Phase error: 23.79 / Stereochemistry target values: ML SAD / Resolution: 2.601→45.785 Å / SU ML: 0.28 / Cross valid method: FREE R-VALUE / σ(F): 1.93 / Phase error: 23.79 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.601→45.785 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj




