+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4ofp | ||||||
|---|---|---|---|---|---|---|---|
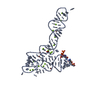
| Title | Crystal Structure of SYG-2 D3-D4 | ||||||
 Components Components | Protein SYG-2 | ||||||
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  Immunoglobulin superfamily / Immunoglobulin superfamily /  Synaptogenesis / Synaptogenesis /  Protein Binding / Protein Binding /  N-linked Glycosylation / N-linked Glycosylation /  Membrane / Membrane /  Extracellular / Extracellular /  SIGNALING PROTEIN SIGNALING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationNephrin family interactions / synaptic target recognition / protein complex involved in cell adhesion / collateral sprouting / protein localization to synapse /  synapse assembly / synapse assembly /  cell adhesion molecule binding / cell adhesion molecule binding /  synaptic membrane / synapse organization / synaptic membrane / synapse organization /  cell-cell adhesion ...Nephrin family interactions / synaptic target recognition / protein complex involved in cell adhesion / collateral sprouting / protein localization to synapse / cell-cell adhesion ...Nephrin family interactions / synaptic target recognition / protein complex involved in cell adhesion / collateral sprouting / protein localization to synapse /  synapse assembly / synapse assembly /  cell adhesion molecule binding / cell adhesion molecule binding /  synaptic membrane / synapse organization / synaptic membrane / synapse organization /  cell-cell adhesion / cell-cell junction / cell-cell signaling / protein homodimerization activity / cell-cell adhesion / cell-cell junction / cell-cell signaling / protein homodimerization activity /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å MOLECULAR REPLACEMENT / Resolution: 3 Å | ||||||
 Authors Authors | Ozkan, E. / Garcia, K.C. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 2014 Journal: Cell(Cambridge,Mass.) / Year: 2014Title: Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis. Authors: Ozkan, E. / Chia, P.H. / Wang, R.R. / Goriatcheva, N. / Borek, D. / Otwinowski, Z. / Walz, T. / Shen, K. / Garcia, K.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4ofp.cif.gz 4ofp.cif.gz | 323.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4ofp.ent.gz pdb4ofp.ent.gz | 265.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4ofp.json.gz 4ofp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/of/4ofp https://data.pdbj.org/pub/pdb/validation_reports/of/4ofp ftp://data.pdbj.org/pub/pdb/validation_reports/of/4ofp ftp://data.pdbj.org/pub/pdb/validation_reports/of/4ofp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4of0C  4of3C  4of6C  4of7C  4of8C  4ofdC  4ofiC  4ofkSC  4ofyC C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 22959.475 Da / Num. of mol.: 4 / Fragment: D3-D4, UNP residues 231-430 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Caenorhabditis elegans (invertebrata) / Gene: C26G2.1, CELE_C26G2.1, syg-2 / Production host: Caenorhabditis elegans (invertebrata) / Gene: C26G2.1, CELE_C26G2.1, syg-2 / Production host:   Trichoplusia ni (cabbage looper) / Strain (production host): High Five / References: UniProt: Q9U3P2 Trichoplusia ni (cabbage looper) / Strain (production host): High Five / References: UniProt: Q9U3P2#2: Sugar | ChemComp-NAG /  N-Acetylglucosamine N-Acetylglucosamine |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.56 Å3/Da / Density % sol: 65.49 % |
|---|---|
Crystal grow | Temperature: 295 K / Method: vapor diffusion, sitting drop / pH: 4.5 Details: 0.9 M Diammonium tartrate, 0.1 M Sodium acetate, pH 4.5, VAPOR DIFFUSION, SITTING DROP, temperature 295.0K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-B / Wavelength: 1.0332 Å / Beamline: 23-ID-B / Wavelength: 1.0332 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Jul 23, 2012 |
| Radiation | Monochromator: Double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.0332 Å / Relative weight: 1 : 1.0332 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. all: 27368 / Num. obs: 27351 / % possible obs: 99.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 18.4 % / Biso Wilson estimate: 106.06 Å2 / Rsym value: 0.158 / Net I/σ(I): 13.15 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB Entry 4OFK Resolution: 3→46.315 Å / SU ML: 0.43 / σ(F): 0 / Phase error: 30.56 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→46.315 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller







 PDBj
PDBj










