+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3owz | ||||||
|---|---|---|---|---|---|---|---|
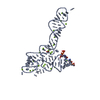
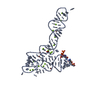
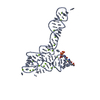
| Title | Crystal structure of glycine riboswitch, soaked in Iridium | ||||||
 Components Components | (Domain II of glycine ...) x 2 | ||||||
 Keywords Keywords |  RNA / RNA /  gene expression / gene expression /  glycine riboswitch glycine riboswitch | ||||||
| Function / homology |  GLYCINE / IRIDIUM HEXAMMINE ION / : / GLYCINE / IRIDIUM HEXAMMINE ION / : /  RNA / RNA (> 10) RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.949 Å MAD / Resolution: 2.949 Å | ||||||
 Authors Authors | Huang, L. / Serganov, A. / Patel, D.J. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2010 Journal: Mol.Cell / Year: 2010Title: Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Authors: Huang, L. / Serganov, A. / Patel, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3owz.cif.gz 3owz.cif.gz | 114.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3owz.ent.gz pdb3owz.ent.gz | 86.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3owz.json.gz 3owz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ow/3owz https://data.pdbj.org/pub/pdb/validation_reports/ow/3owz ftp://data.pdbj.org/pub/pdb/validation_reports/ow/3owz ftp://data.pdbj.org/pub/pdb/validation_reports/ow/3owz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3owiC  3owwC  3ox0C  3oxbC  3oxdC  3oxeC  3oxjC  3oxmC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| Unit cell |
|
- Components
Components
-Domain II of glycine ... , 2 types, 2 molecules AB
| #1: RNA chain | Mass: 28735.090 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: in vitro transcription / References: GenBank: CP001485.1 |
|---|---|
| #2: RNA chain | Mass: 28655.109 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: in vitro transcription / References: GenBank: CP001485.1 |
-Non-polymers , 4 types, 112 molecules 





| #3: Chemical |  Glycine Glycine#4: Chemical | ChemComp-MG / #5: Chemical | ChemComp-IRI / #6: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Nonpolymer details | THE ORIGINAL MG AND IRI CHAIN IDS HAVE BEEN CHANGED TO THE CLOSEST POLYMER CHAIN. 100 HAS BEEN ...THE ORIGINAL MG AND IRI CHAIN IDS HAVE BEEN CHANGED TO THE CLOSEST POLYMER CHAIN. 100 HAS BEEN ADDED TO THE ORIGINAL MG RESIDUE NUMBER AND 200 TO THE ORIGINAL IRI NUMBER. |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.57 Å3/Da / Density % sol: 52.08 % |
|---|---|
Crystal grow | Temperature: 293.15 K / Method: vapor diffusion, hanging drop / pH: 5.1 Details: 0.05 M Na-cacodylate, pH 5.1, 0.2 M KCl, 8 % (w/v) PEG8000 and 80 mM magnesium acetate, VAPOR DIFFUSION, HANGING DROP, temperature 293.15K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X29A / Wavelength: 1.1053,1.1056,1.1256,1.10859 / Beamline: X29A / Wavelength: 1.1053,1.1056,1.1256,1.10859 | |||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Jun 10, 2009 | |||||||||||||||
| Radiation | Monochromator: SI mirror / Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Resolution: 2.949→20 Å / Num. all: 12933 / Num. obs: 12921 / % possible obs: 99.9 % / Observed criterion σ(F): -3 / Observed criterion σ(I): -3 / Redundancy: 6.9 % / Rmerge(I) obs: 0.09 / Net I/σ(I): 38 | |||||||||||||||
| Reflection shell | Resolution: 2.949→3 Å / Redundancy: 7.1 % / Rmerge(I) obs: 0.385 / % possible all: 100 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MAD / Resolution: 2.949→20 Å / Cor.coef. Fo:Fc: 0.927 / Cor.coef. Fo:Fc free: 0.906 / SU B: 14.223 / SU ML: 0.272 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.418 / Stereochemistry target values: MAXIMUM LIKELIHOOD MAD / Resolution: 2.949→20 Å / Cor.coef. Fo:Fc: 0.927 / Cor.coef. Fo:Fc free: 0.906 / SU B: 14.223 / SU ML: 0.272 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.418 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| |||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.949→20 Å
| |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.949→3.026 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller










 PDBj
PDBj


































