+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4nce | ||||||
|---|---|---|---|---|---|---|---|
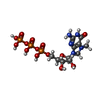
| Title | Influenza polymerase basic protein 2 (PB2) bound to 7-methyl-GTP | ||||||
 Components Components | Polymerase basic protein 2 | ||||||
 Keywords Keywords | TRANSCRIPTION/TRANSCRIPTION INHIBITOR /  influenza virus / cap-binding domain / TRANSCRIPTION-TRANSCRIPTION INHIBITOR complex influenza virus / cap-binding domain / TRANSCRIPTION-TRANSCRIPTION INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information cap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / cap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity /  virion component / virus-mediated perturbation of host defense response / viral RNA genome replication / virion component / virus-mediated perturbation of host defense response / viral RNA genome replication /  RNA-dependent RNA polymerase activity / DNA-templated transcription ... RNA-dependent RNA polymerase activity / DNA-templated transcription ... cap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / cap snatching / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / host cell mitochondrion / 7-methylguanosine mRNA capping / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity /  virion component / virus-mediated perturbation of host defense response / viral RNA genome replication / virion component / virus-mediated perturbation of host defense response / viral RNA genome replication /  RNA-dependent RNA polymerase activity / DNA-templated transcription / host cell nucleus / RNA-dependent RNA polymerase activity / DNA-templated transcription / host cell nucleus /  RNA binding RNA bindingSimilarity search - Function | ||||||
| Biological species |    Influenza A virus Influenza A virus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | ||||||
 Authors Authors | Jacobs, M.D. | ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2014 Journal: J.Med.Chem. / Year: 2014Title: Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2. Authors: Clark, M.P. / Ledeboer, M.W. / Davies, I. / Byrn, R.A. / Jones, S.M. / Perola, E. / Tsai, A. / Jacobs, M. / Nti-Addae, K. / Bandarage, U.K. / Boyd, M.J. / Bethiel, R.S. / Court, J.J. / Deng, ...Authors: Clark, M.P. / Ledeboer, M.W. / Davies, I. / Byrn, R.A. / Jones, S.M. / Perola, E. / Tsai, A. / Jacobs, M. / Nti-Addae, K. / Bandarage, U.K. / Boyd, M.J. / Bethiel, R.S. / Court, J.J. / Deng, H. / Duffy, J.P. / Dorsch, W.A. / Farmer, L.J. / Gao, H. / Gu, W. / Jackson, K. / Jacobs, D.H. / Kennedy, J.M. / Ledford, B. / Liang, J. / Maltais, F. / Murcko, M. / Wang, T. / Wannamaker, M.W. / Bennett, H.B. / Leeman, J.R. / McNeil, C. / Taylor, W.P. / Memmott, C. / Jiang, M. / Rijnbrand, R. / Bral, C. / Germann, U. / Nezami, A. / Zhang, Y. / Salituro, F.G. / Bennani, Y.L. / Charifson, P.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4nce.cif.gz 4nce.cif.gz | 83.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4nce.ent.gz pdb4nce.ent.gz | 62 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4nce.json.gz 4nce.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nc/4nce https://data.pdbj.org/pub/pdb/validation_reports/nc/4nce ftp://data.pdbj.org/pub/pdb/validation_reports/nc/4nce ftp://data.pdbj.org/pub/pdb/validation_reports/nc/4nce | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4ncmC  4p1uC  2vqzS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 19174.324 Da / Num. of mol.: 1 / Fragment: CAP-BINDING DOMAIN / Mutation: R389K Source method: isolated from a genetically manipulated source Source: (gene. exp.)    Influenza A virus / Strain: strain A/Victoria/3/1975 H3N2 / Gene: PB2, polymerase basic protein / Plasmid details: with an N-terminal TEV cleavable 6*His tag / Plasmid: pET28b / Production host: Influenza A virus / Strain: strain A/Victoria/3/1975 H3N2 / Gene: PB2, polymerase basic protein / Plasmid details: with an N-terminal TEV cleavable 6*His tag / Plasmid: pET28b / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) RIPL / References: UniProt: P31345 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) RIPL / References: UniProt: P31345 | ||
|---|---|---|---|
| #2: Chemical | ChemComp-MGT / | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.72 Å3/Da / Density % sol: 54.85 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 4.7 Details: 0.7 uL of protein solution (9.7 mg/mL protein, 50 mM Tris buffer pH 8, 200 mM sodium chloride, 2 mM dithiothreitol, 5 mM m7GTP) was mixed with 0.35 uL Hampton Research Silver Bullet ...Details: 0.7 uL of protein solution (9.7 mg/mL protein, 50 mM Tris buffer pH 8, 200 mM sodium chloride, 2 mM dithiothreitol, 5 mM m7GTP) was mixed with 0.35 uL Hampton Research Silver Bullet screening reagent #36. This mixture was combined with 0.35 uL of well solution (approximately 1.3 M sodium formate, 100 mM sodium citrate buffer pH 4.6, 5 mM dithiothreitol) and suspended over 1 mL of well solution, VAPOR DIFFUSION, HANGING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1 Å / Beamline: 5.0.2 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Aug 19, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→29.62 Å / Num. all: 9272 / Num. obs: 9272 / % possible obs: 100 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 1 / Redundancy: 6.45 % / Rmerge(I) obs: 0.134 / Net I/σ(I): 8.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2VQZ Resolution: 2.3→29.62 Å / Cor.coef. Fo:Fc: 0.942 / Cor.coef. Fo:Fc free: 0.916 / SU B: 15.39 / SU ML: 0.167 / Isotropic thermal model: Isotropic / Cross valid method: THROUGHOUT / ESU R: 0.31 / ESU R Free: 0.216 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.577 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→29.62 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.36 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj