[English] 日本語
 Yorodumi
Yorodumi- PDB-4n11: Crystal structure of plasmodium falciparum reduced glutaredoxin 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4n11 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of plasmodium falciparum reduced glutaredoxin 1 (PfGrx1) complexed with cisplatin | ||||||
 Components Components | Glutaredoxin | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  Glutathione / Glutathione /  Active site / TRX FOLD / Redox Enzyme / Pt-SAD / Active site / TRX FOLD / Redox Enzyme / Pt-SAD /  cisplatin cisplatin | ||||||
| Function / homology |  Function and homology information Function and homology information protein-disulfide reductase (glutathione) activity / Interconversion of nucleotide di- and triphosphates / glutathione disulfide oxidoreductase activity / protein-disulfide reductase (glutathione) activity / Interconversion of nucleotide di- and triphosphates / glutathione disulfide oxidoreductase activity /  antioxidant activity antioxidant activitySimilarity search - Function | ||||||
| Biological species |   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SAD / Resolution: 1.97 Å SAD / Resolution: 1.97 Å | ||||||
 Authors Authors | Yogavel, M. / Sharma, A. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Interaction of Cisplatin with plasmodium falciparum Glutaredoxin 1 Authors: Yogavel, M. / Tripathi, T. / Rahlfs, S. / Becker, K. / Sharma, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4n11.cif.gz 4n11.cif.gz | 39.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4n11.ent.gz pdb4n11.ent.gz | 26.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4n11.json.gz 4n11.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/n1/4n11 https://data.pdbj.org/pub/pdb/validation_reports/n1/4n11 ftp://data.pdbj.org/pub/pdb/validation_reports/n1/4n11 ftp://data.pdbj.org/pub/pdb/validation_reports/n1/4n11 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / Glutaredoxin 1 / Glutaredoxin 1Mass: 12436.457 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum)Strain: 3D7 / Gene: GRX1 / Plasmid: pQE30 / Production host:   Escherichia Coli (E. coli) / Strain (production host): M15 Escherichia Coli (E. coli) / Strain (production host): M15References: UniProt: Q9NLB2, arsenate reductase (glutathione/glutaredoxin) | ||||
|---|---|---|---|---|---|
| #2: Chemical | ChemComp-MPO /  MOPS MOPS | ||||
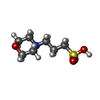
| #3: Chemical |  2-Methyl-2,4-pentanediol 2-Methyl-2,4-pentanediol#4: Chemical | ChemComp-CPT / |  Cisplatin Cisplatin#5: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.25 Å3/Da / Density % sol: 45.27 % / Mosaicity: 1.599 ° |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7.5 Details: 12.5%(W/V) PEG1000, 12.5%(W/V) PEG3350, 12.5%(V/V) MPD, 0.02M AMINO ACIDS, 0.1M MOPS/HEPES SODIUM,, pH 7.5, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.5418 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RAYONIX MX-225 / Detector: CCD / Date: Jul 17, 2013 / Details: mirrors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: SAGITALLY FOCUSED Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.97→50 Å / Num. all: 8380 / Num. obs: 8372 / % possible obs: 99.9 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 2 / Redundancy: 10 % / Biso Wilson estimate: 27.99 Å2 / Rmerge(I) obs: 0.075 / Χ2: 1.247 / Net I/σ(I): 10.6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 1.97→27.464 Å / Occupancy max: 1 / Occupancy min: 0.3 / FOM work R set: 0.8812 / SU ML: 0.16 / σ(F): 1.36 / Phase error: 18.03 / Stereochemistry target values: ML SAD / Resolution: 1.97→27.464 Å / Occupancy max: 1 / Occupancy min: 0.3 / FOM work R set: 0.8812 / SU ML: 0.16 / σ(F): 1.36 / Phase error: 18.03 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 75.7 Å2 / Biso mean: 27.4628 Å2 / Biso min: 15.33 Å2 | ||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.97→27.464 Å
| ||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 6 / % reflection obs: 100 %
|
 Movie
Movie Controller
Controller


















 PDBj
PDBj