+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4cim | ||||||
|---|---|---|---|---|---|---|---|
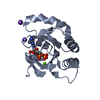
| Title | Complex of a Bcl-w BH3 mutant with a BH3 domain | ||||||
 Components Components | (BCL-2-LIKE PROTEIN 2) x 2 | ||||||
 Keywords Keywords |  APOPTOSIS / APOPTOSIS /  CELL DEATH CELL DEATH | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mitochondrial membrane permeability / Sertoli cell proliferation /  BH domain binding / BH domain binding /  Bcl-2 family protein complex / negative regulation of release of cytochrome c from mitochondria / cellular response to glycine / negative regulation of intrinsic apoptotic signaling pathway / extrinsic apoptotic signaling pathway in absence of ligand / cellular response to estradiol stimulus / response to ischemia ...negative regulation of mitochondrial membrane permeability / Sertoli cell proliferation / Bcl-2 family protein complex / negative regulation of release of cytochrome c from mitochondria / cellular response to glycine / negative regulation of intrinsic apoptotic signaling pathway / extrinsic apoptotic signaling pathway in absence of ligand / cellular response to estradiol stimulus / response to ischemia ...negative regulation of mitochondrial membrane permeability / Sertoli cell proliferation /  BH domain binding / BH domain binding /  Bcl-2 family protein complex / negative regulation of release of cytochrome c from mitochondria / cellular response to glycine / negative regulation of intrinsic apoptotic signaling pathway / extrinsic apoptotic signaling pathway in absence of ligand / cellular response to estradiol stimulus / response to ischemia / cellular response to amyloid-beta / intrinsic apoptotic signaling pathway in response to DNA damage / disordered domain specific binding / Bcl-2 family protein complex / negative regulation of release of cytochrome c from mitochondria / cellular response to glycine / negative regulation of intrinsic apoptotic signaling pathway / extrinsic apoptotic signaling pathway in absence of ligand / cellular response to estradiol stimulus / response to ischemia / cellular response to amyloid-beta / intrinsic apoptotic signaling pathway in response to DNA damage / disordered domain specific binding /  spermatogenesis / regulation of apoptotic process / mitochondrial outer membrane / protein heterodimerization activity / protein-containing complex binding / negative regulation of apoptotic process / protein homodimerization activity / identical protein binding / spermatogenesis / regulation of apoptotic process / mitochondrial outer membrane / protein heterodimerization activity / protein-containing complex binding / negative regulation of apoptotic process / protein homodimerization activity / identical protein binding /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||
 Authors Authors | Colman, P.M. / Lee, E.F. / Fairlie, W.D. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2014 Journal: J.Biol.Chem. / Year: 2014Title: The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains Authors: Lee, E.F. / Dewson, G. / Evangelista, M. / Pettikiriarachchi, A. / Zhu, H. / Colman, P.M. / Fairlie, W.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4cim.cif.gz 4cim.cif.gz | 149.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4cim.ent.gz pdb4cim.ent.gz | 118.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4cim.json.gz 4cim.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ci/4cim https://data.pdbj.org/pub/pdb/validation_reports/ci/4cim ftp://data.pdbj.org/pub/pdb/validation_reports/ci/4cim ftp://data.pdbj.org/pub/pdb/validation_reports/ci/4cim | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4cinC  2y6wS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 17801.867 Da / Num. of mol.: 2 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92843 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92843#2: Protein/peptide | Mass: 2336.583 Da / Num. of mol.: 2 / Fragment: RESIDUES 38-58 / Mutation: YES Source method: isolated from a genetically manipulated source Details: PEPTIDE FROM PROTEOLYTIC CLEAVAGE BCL-W DURING PURIFICATION Source: (gene. exp.)   HOMO SAPIENS (human) / Production host: HOMO SAPIENS (human) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92843 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q92843#3: Chemical | ChemComp-EDO /  Ethylene glycol Ethylene glycol#4: Water | ChemComp-HOH / |  Water WaterSequence details | MUTATIONS IN SEQUENCE AS FOLLOWS C29S H43G M46L A49I A128E C-TERMINAL TRUNCATION | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.04 Å3/Da / Density % sol: 39.7 % / Description: NONE |
|---|---|
Crystal grow | pH: 8 / Details: 1 M TRISODIUM CITRATE 0.1 M IMIDAZOLE, PH 8.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.954 / Beamline: MX2 / Wavelength: 0.954 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.954 Å / Relative weight: 1 : 0.954 Å / Relative weight: 1 |
| Reflection | Resolution: 1.5→50 Å / Num. obs: 51625 / % possible obs: 99.4 % / Observed criterion σ(I): 2 / Redundancy: 3.78 % / Rmerge(I) obs: 0.07 / Net I/σ(I): 9.93 |
| Reflection shell | Resolution: 1.5→1.59 Å / Redundancy: 3.64 % / Rmerge(I) obs: 0.85 / Mean I/σ(I) obs: 1.57 / % possible all: 96.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2Y6W Resolution: 1.5→44.229 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→44.229 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



