+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4b2z | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of Osh6 in complex with phosphatidylserine | ||||||
 Components Components | Oxysterol-binding protein homolog 6 | ||||||
 Keywords Keywords |  TRANSPORT PROTEIN / LIPID TRANSPORT TRANSPORT PROTEIN / LIPID TRANSPORT | ||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis of bile acids and bile salts / Acyl chain remodelling of PS / sterol transfer activity / : / sterol transport / sterol homeostasis / phospholipid transporter activity /  sterol binding / cortical endoplasmic reticulum / phosphatidylinositol-5-phosphate binding ...Synthesis of bile acids and bile salts / Acyl chain remodelling of PS / sterol transfer activity / : / sterol transport / sterol homeostasis / phospholipid transporter activity / sterol binding / cortical endoplasmic reticulum / phosphatidylinositol-5-phosphate binding ...Synthesis of bile acids and bile salts / Acyl chain remodelling of PS / sterol transfer activity / : / sterol transport / sterol homeostasis / phospholipid transporter activity /  sterol binding / cortical endoplasmic reticulum / phosphatidylinositol-5-phosphate binding / sterol metabolic process / maintenance of cell polarity / piecemeal microautophagy of the nucleus / sterol binding / cortical endoplasmic reticulum / phosphatidylinositol-5-phosphate binding / sterol metabolic process / maintenance of cell polarity / piecemeal microautophagy of the nucleus /  phosphatidic acid binding / phospholipid transport / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylinositol-4-phosphate binding / phosphatidylinositol-3,5-bisphosphate binding / phosphatidic acid binding / phospholipid transport / phosphatidylinositol-3,4-bisphosphate binding / phosphatidylinositol-4-phosphate binding / phosphatidylinositol-3,5-bisphosphate binding /  phosphatidylserine binding / phosphatidylserine binding /  exocytosis / exocytosis /  endocytosis / intracellular membrane-bounded organelle / endocytosis / intracellular membrane-bounded organelle /  lipid binding / endoplasmic reticulum membrane / lipid binding / endoplasmic reticulum membrane /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å MOLECULAR REPLACEMENT / Resolution: 1.95 Å | ||||||
 Authors Authors | Maeda, K. / Anand, K. / Chiapparino, A. / Kumar, A. / Poletto, M. / Kaksonen, M. / Gavin, A.C. | ||||||
 Citation Citation |  Journal: Nature / Year: 2013 Journal: Nature / Year: 2013Title: Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins Authors: Maeda, K. / Anand, K. / Chiapparino, A. / Kumar, A. / Poletto, M. / Kaksonen, M. / Gavin, A.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4b2z.cif.gz 4b2z.cif.gz | 355.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4b2z.ent.gz pdb4b2z.ent.gz | 289.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4b2z.json.gz 4b2z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b2/4b2z https://data.pdbj.org/pub/pdb/validation_reports/b2/4b2z ftp://data.pdbj.org/pub/pdb/validation_reports/b2/4b2z ftp://data.pdbj.org/pub/pdb/validation_reports/b2/4b2z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1zhtS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||
| 2 | 
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 51664.891 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Gene: OSH6, YKR003W, YK102 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Star / References: UniProt: Q02201 Escherichia coli BL21(DE3) (bacteria) / Variant (production host): Star / References: UniProt: Q02201 |
|---|
-Non-polymers , 5 types, 369 molecules 

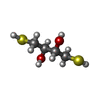






| #2: Chemical |  Sulfate Sulfate#3: Chemical |  Phosphatidylserine Phosphatidylserine#4: Chemical | ChemComp-DTU / ( |  Dithioerythritol Dithioerythritol#5: Chemical | ChemComp-DTT / |  Dithiothreitol Dithiothreitol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 5 X-RAY DIFFRACTION / Number of used crystals: 5 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 49.2 % / Description: NONE |
|---|---|
Crystal grow | pH: 6.5 / Details: 0.1M MES PH6.5, 13% PEG6000, 5% MPD |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-1 / Wavelength: 0.9334 / Beamline: ID14-1 / Wavelength: 0.9334 |
| Detector | Type: ADSC QUANTUM 210 / Detector: CCD / Date: Jun 16, 2011 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9334 Å / Relative weight: 1 : 0.9334 Å / Relative weight: 1 |
| Reflection | Resolution: 1.95→40 Å / Num. obs: 65896 / % possible obs: 91.8 % / Observed criterion σ(I): 1.63 / Redundancy: 3.8 % / Biso Wilson estimate: 25.9 Å2 / Rmerge(I) obs: 0.06 / Net I/σ(I): 18 |
| Reflection shell | Resolution: 1.95→2 Å / Redundancy: 2.8 % / Rmerge(I) obs: 0.65 / Mean I/σ(I) obs: 1.6 / % possible all: 59 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1ZHT Resolution: 1.95→46.566 Å / SU ML: 0.26 / σ(F): 1.99 / Phase error: 24.76 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 31.39 Å2 / ksol: 0.343 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 33 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95→46.566 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 10.9179 Å / Origin y: -17.9992 Å / Origin z: 30.9626 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: ALL |
 Movie
Movie Controller
Controller












 PDBj
PDBj







