+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3vzc | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Sphingosine Kinase 1 with inhibitor | ||||||
 Components Components | Sphingosine kinase 1 | ||||||
 Keywords Keywords | TRANSFERASE/INHIBITOR /  lipid kinase / TRANSFERASE-INHIBITOR complex lipid kinase / TRANSFERASE-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information sphingosine kinase / sphingosine kinase /  sphinganine kinase activity / D-erythro-sphingosine kinase activity / sphingoid catabolic process / regulation of endosomal vesicle fusion / sphingosine metabolic process / negative regulation of ceramide biosynthetic process / sphinganine kinase activity / D-erythro-sphingosine kinase activity / sphingoid catabolic process / regulation of endosomal vesicle fusion / sphingosine metabolic process / negative regulation of ceramide biosynthetic process /  sphingosine-1-phosphate receptor activity / regulation of microglial cell activation / regulation of interleukin-1 beta production ... sphingosine-1-phosphate receptor activity / regulation of microglial cell activation / regulation of interleukin-1 beta production ... sphingosine kinase / sphingosine kinase /  sphinganine kinase activity / D-erythro-sphingosine kinase activity / sphingoid catabolic process / regulation of endosomal vesicle fusion / sphingosine metabolic process / negative regulation of ceramide biosynthetic process / sphinganine kinase activity / D-erythro-sphingosine kinase activity / sphingoid catabolic process / regulation of endosomal vesicle fusion / sphingosine metabolic process / negative regulation of ceramide biosynthetic process /  sphingosine-1-phosphate receptor activity / regulation of microglial cell activation / regulation of interleukin-1 beta production / sphingosine biosynthetic process / sphingolipid biosynthetic process / regulation of neuroinflammatory response / Sphingolipid de novo biosynthesis / positive regulation of smooth muscle contraction / sphingosine-1-phosphate receptor activity / regulation of microglial cell activation / regulation of interleukin-1 beta production / sphingosine biosynthetic process / sphingolipid biosynthetic process / regulation of neuroinflammatory response / Sphingolipid de novo biosynthesis / positive regulation of smooth muscle contraction /  regulation of phagocytosis / regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of p38MAPK cascade / DNA biosynthetic process / regulation of phagocytosis / regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of p38MAPK cascade / DNA biosynthetic process /  protein acetylation / blood vessel development / positive regulation of interleukin-17 production / Association of TriC/CCT with target proteins during biosynthesis / protein acetylation / blood vessel development / positive regulation of interleukin-17 production / Association of TriC/CCT with target proteins during biosynthesis /  acetyltransferase activity / acetyltransferase activity /  regulation of endocytosis / endocytic vesicle / cellular response to vascular endothelial growth factor stimulus / response to tumor necrosis factor / regulation of endocytosis / endocytic vesicle / cellular response to vascular endothelial growth factor stimulus / response to tumor necrosis factor /  clathrin-coated pit / clathrin-coated pit /  Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / positive regulation of mitotic cell cycle / positive regulation of mitotic nuclear division / protein phosphatase 2A binding / positive regulation of peptidyl-threonine phosphorylation / VEGFR2 mediated cell proliferation / positive regulation of protein ubiquitination / calcium-mediated signaling / Transferases; Acyltransferases; Transferring groups other than aminoacyl groups / positive regulation of mitotic cell cycle / positive regulation of mitotic nuclear division / protein phosphatase 2A binding / positive regulation of peptidyl-threonine phosphorylation / VEGFR2 mediated cell proliferation / positive regulation of protein ubiquitination / calcium-mediated signaling /  brain development / PKR-mediated signaling / cellular response to growth factor stimulus / cellular response to hydrogen peroxide / positive regulation of non-canonical NF-kappaB signal transduction / positive regulation of angiogenesis / positive regulation of fibroblast proliferation / presynapse / positive regulation of NF-kappaB transcription factor activity / early endosome membrane / positive regulation of cell growth / cell population proliferation / Extra-nuclear estrogen signaling / brain development / PKR-mediated signaling / cellular response to growth factor stimulus / cellular response to hydrogen peroxide / positive regulation of non-canonical NF-kappaB signal transduction / positive regulation of angiogenesis / positive regulation of fibroblast proliferation / presynapse / positive regulation of NF-kappaB transcription factor activity / early endosome membrane / positive regulation of cell growth / cell population proliferation / Extra-nuclear estrogen signaling /  calmodulin binding / intracellular signal transduction / positive regulation of cell migration / calmodulin binding / intracellular signal transduction / positive regulation of cell migration /  inflammatory response / inflammatory response /  phosphorylation / intracellular membrane-bounded organelle / phosphorylation / intracellular membrane-bounded organelle /  lipid binding / negative regulation of apoptotic process / magnesium ion binding / lipid binding / negative regulation of apoptotic process / magnesium ion binding /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  membrane / membrane /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   homo sapiens (human) homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.3 Å MAD / Resolution: 2.3 Å | ||||||
 Authors Authors | Min, X. / Walker, N.P. / Wang, Z. | ||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Authors: Wang, Z. / Min, X. / Xiao, S.H. / Johnstone, S. / Romanow, W. / Meininger, D. / Xu, H. / Liu, J. / Dai, J. / An, S. / Thibault, S. / Walker, N. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3vzc.cif.gz 3vzc.cif.gz | 407.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3vzc.ent.gz pdb3vzc.ent.gz | 333.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3vzc.json.gz 3vzc.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vz/3vzc https://data.pdbj.org/pub/pdb/validation_reports/vz/3vzc ftp://data.pdbj.org/pub/pdb/validation_reports/vz/3vzc ftp://data.pdbj.org/pub/pdb/validation_reports/vz/3vzc | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| Unit cell |
|
- Components
Components
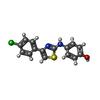
| #1: Protein |  / SK 1 / SPK 1 / SK 1 / SPK 1Mass: 39917.469 Da / Num. of mol.: 6 / Fragment: UNP RESIDUES 9-364 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   homo sapiens (human) / Cell line: sf9 / Gene: SPHK1, SPHK, SPK / Plasmid: pFastBac HTb / References: UniProt: Q9NYA1, homo sapiens (human) / Cell line: sf9 / Gene: SPHK1, SPHK, SPK / Plasmid: pFastBac HTb / References: UniProt: Q9NYA1,  sphingosine kinase sphingosine kinase#2: Chemical | ChemComp-UUL / #3: Chemical |  Ethylene glycol Ethylene glycol#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.57 Å3/Da / Density % sol: 52.16 % / Mosaicity: 0.34 ° |
|---|---|
Crystal grow | Temperature: 289 K / Method: sitting drop / pH: 6 Details: 1.0-1.4M ammonium sulfate, 0.3-1.3M NaCl, 0.1M Bis-Tris, pH 6.0, sitting drop, temperature 289K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1 Å / Beamline: 5.0.2 / Wavelength: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC315R / Detector: CCD / Details: 3x3 CCD array | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Double-crystal, Si(111) / Protocol: MAD / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.3→113.09 Å / Num. all: 101561 / Num. obs: 101561 / % possible obs: 92.1 % / Redundancy: 3.4 % / Rsym value: 0.057 / Net I/σ(I): 12.6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MAD / Resolution: 2.3→50 Å / Cor.coef. Fo:Fc: 0.944 / Cor.coef. Fo:Fc free: 0.911 / WRfactor Rfree: 0.2877 / WRfactor Rwork: 0.2323 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.7464 / SU B: 8.889 / SU ML: 0.213 / SU R Cruickshank DPI: 0.4048 / SU Rfree: 0.283 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.405 / ESU R Free: 0.283 / Stereochemistry target values: MAXIMUM LIKELIHOOD MAD / Resolution: 2.3→50 Å / Cor.coef. Fo:Fc: 0.944 / Cor.coef. Fo:Fc free: 0.911 / WRfactor Rfree: 0.2877 / WRfactor Rwork: 0.2323 / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.7464 / SU B: 8.889 / SU ML: 0.213 / SU R Cruickshank DPI: 0.4048 / SU Rfree: 0.283 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.405 / ESU R Free: 0.283 / Stereochemistry target values: MAXIMUM LIKELIHOODDetails: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT U VALUES: REFINED INDIVIDUALLY
| |||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 127.7 Å2 / Biso mean: 44.2649 Å2 / Biso min: 16.49 Å2
| |||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→50 Å
| |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.36 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller















 PDBj
PDBj