[English] 日本語
 Yorodumi
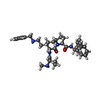
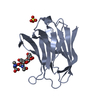
Yorodumi- PDB-3oz1: cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac066 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3oz1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac066 | ||||||
 Components Components | Baculoviral IAP repeat-containing protein 2 | ||||||
 Keywords Keywords | APOPTOSIS INHIBITOR /  Zinc-finger / Caspases Zinc-finger / Caspases | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of ripoptosome assembly involved in necroptotic process / regulation of cysteine-type endopeptidase activity / FBXO family protein binding / regulation of RIG-I signaling pathway / positive regulation of protein K48-linked ubiquitination / regulation of non-canonical NF-kappaB signal transduction / TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway / regulation of necroptotic process / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / positive regulation of protein K63-linked ubiquitination ...negative regulation of ripoptosome assembly involved in necroptotic process / regulation of cysteine-type endopeptidase activity / FBXO family protein binding / regulation of RIG-I signaling pathway / positive regulation of protein K48-linked ubiquitination / regulation of non-canonical NF-kappaB signal transduction / TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway / regulation of necroptotic process / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / positive regulation of protein K63-linked ubiquitination / CD40 receptor complex / XY body / negative regulation of necroptotic process / positive regulation of protein monoubiquitination / regulation of reactive oxygen species metabolic process / TNFR1-induced proapoptotic signaling / RIPK1-mediated regulated necrosis /  regulation of toll-like receptor signaling pathway / regulation of toll-like receptor signaling pathway /  regulation of innate immune response / Apoptotic cleavage of cellular proteins / regulation of innate immune response / Apoptotic cleavage of cellular proteins /  regulation of cell differentiation / non-canonical NF-kappaB signal transduction / necroptotic process / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / canonical NF-kappaB signal transduction / response to cAMP / tumor necrosis factor-mediated signaling pathway / TICAM1, RIP1-mediated IKK complex recruitment / IKK complex recruitment mediated by RIP1 / placenta development / TNFR1-induced NF-kappa-B signaling pathway / regulation of cell differentiation / non-canonical NF-kappaB signal transduction / necroptotic process / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / canonical NF-kappaB signal transduction / response to cAMP / tumor necrosis factor-mediated signaling pathway / TICAM1, RIP1-mediated IKK complex recruitment / IKK complex recruitment mediated by RIP1 / placenta development / TNFR1-induced NF-kappa-B signaling pathway /  ubiquitin binding / positive regulation of protein ubiquitination / TNFR2 non-canonical NF-kB pathway / Regulation of TNFR1 signaling / RING-type E3 ubiquitin transferase / NOD1/2 Signaling Pathway / Regulation of necroptotic cell death / cytoplasmic side of plasma membrane / protein polyubiquitination / ubiquitin-protein transferase activity / ubiquitin binding / positive regulation of protein ubiquitination / TNFR2 non-canonical NF-kB pathway / Regulation of TNFR1 signaling / RING-type E3 ubiquitin transferase / NOD1/2 Signaling Pathway / Regulation of necroptotic cell death / cytoplasmic side of plasma membrane / protein polyubiquitination / ubiquitin-protein transferase activity /  ubiquitin protein ligase activity / regulation of cell population proliferation / ubiquitin protein ligase activity / regulation of cell population proliferation /  regulation of inflammatory response / protein-folding chaperone binding / regulation of inflammatory response / protein-folding chaperone binding /  transferase activity / proteasome-mediated ubiquitin-dependent protein catabolic process / response to ethanol / regulation of apoptotic process / positive regulation of canonical NF-kappaB signal transduction / transferase activity / proteasome-mediated ubiquitin-dependent protein catabolic process / response to ethanol / regulation of apoptotic process / positive regulation of canonical NF-kappaB signal transduction /  transcription coactivator activity / response to hypoxia / cell surface receptor signaling pathway / transcription coactivator activity / response to hypoxia / cell surface receptor signaling pathway /  regulation of cell cycle / Ub-specific processing proteases / apoptotic process / protein-containing complex binding / negative regulation of apoptotic process / zinc ion binding / identical protein binding / regulation of cell cycle / Ub-specific processing proteases / apoptotic process / protein-containing complex binding / negative regulation of apoptotic process / zinc ion binding / identical protein binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å MOLECULAR REPLACEMENT / Resolution: 3 Å | ||||||
 Authors Authors | Cossu, F. / Malvezzi, F. / Mastrangelo, E. / Canevari, G. / Bolognesi, M. / Milani, M. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2010 Journal: Protein Sci. / Year: 2010Title: Recognition of Smac-mimetic compounds by the BIR3 domain of cIAP1 Authors: Cossu, F. / Malvezzi, F. / Canevari, G. / Mastrangelo, E. / Lecis, D. / Delia, D. / Seneci, P. / Scolastico, C. / Bolognesi, M. / Milani, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3oz1.cif.gz 3oz1.cif.gz | 195.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3oz1.ent.gz pdb3oz1.ent.gz | 159.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3oz1.json.gz 3oz1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oz/3oz1 https://data.pdbj.org/pub/pdb/validation_reports/oz/3oz1 ftp://data.pdbj.org/pub/pdb/validation_reports/oz/3oz1 ftp://data.pdbj.org/pub/pdb/validation_reports/oz/3oz1 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14390.241 Da / Num. of mol.: 4 / Fragment: Repeat 3 (BIR3) domain, UNP residues 251-363 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: BIRC2, API1, IAP2, MIHB, RNF48 / Plasmid: pET21A(+) / Production host: Homo sapiens (human) / Gene: BIRC2, API1, IAP2, MIHB, RNF48 / Plasmid: pET21A(+) / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) / References: UniProt: Q13490 Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) / References: UniProt: Q13490#2: Chemical | ChemComp-ZN / #3: Chemical | ChemComp-BMB / ( #4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.65 Å3/Da / Density % sol: 53.63 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 25% PEG 400, 0.1M Sodium Citrate, 0.1M Tris, pH 8.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 70 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.94 Å / Beamline: ID14-4 / Wavelength: 0.94 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: May 17, 2010 |
| Radiation | Monochromator: Si (111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.94 Å / Relative weight: 1 : 0.94 Å / Relative weight: 1 |
| Reflection | Resolution: 3→92.9 Å / Num. all: 11909 / Num. obs: 11333 / % possible obs: 95.2 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 1 / Biso Wilson estimate: 78.37 Å2 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 3→60.93 Å / Cor.coef. Fo:Fc: 0.9153 / Cor.coef. Fo:Fc free: 0.8493 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: MAXIMUM LIKELIHOOD MOLECULAR REPLACEMENT / Resolution: 3→60.93 Å / Cor.coef. Fo:Fc: 0.9153 / Cor.coef. Fo:Fc free: 0.8493 / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: MAXIMUM LIKELIHOOD
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 85.03 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.419 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→60.93 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3→3.29 Å / Total num. of bins used: 6
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj