[English] 日本語
 Yorodumi
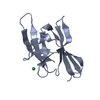
Yorodumi- PDB-2rpi: The NMR structure of the submillisecond folding intermediate of t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2rpi | ||||||
|---|---|---|---|---|---|---|---|
| Title | The NMR structure of the submillisecond folding intermediate of the Thermus thermophilus ribonuclease H | ||||||
 Components Components | Ribonuclease H | ||||||
 Keywords Keywords |  HYDROLASE / submillisecond folding intermediate / Thermus thermophilus ribonuclease H / High-resolution structure / HYDROLASE / submillisecond folding intermediate / Thermus thermophilus ribonuclease H / High-resolution structure /  Endonuclease / Endonuclease /  Magnesium / Metal-binding / Magnesium / Metal-binding /  Nuclease Nuclease | ||||||
| Function / homology |  Function and homology information Function and homology information DNA replication, removal of RNA primer / DNA replication, removal of RNA primer /  ribonuclease H / RNA-DNA hybrid ribonuclease activity / ribonuclease H / RNA-DNA hybrid ribonuclease activity /  nucleic acid binding / magnesium ion binding / nucleic acid binding / magnesium ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |    Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | ||||||
| Method |  SOLUTION NMR / DGSA-distance geometry simulated annealing, torsion angle dynamics SOLUTION NMR / DGSA-distance geometry simulated annealing, torsion angle dynamics | ||||||
 Authors Authors | Zhou, Z. / Feng, H. / Bai, Y. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2008 Journal: J.Mol.Biol. / Year: 2008Title: The high-resolution NMR structure of the early folding intermediate of the Thermus thermophilus ribonuclease H Authors: Zhou, Z. / Feng, H. / Ghirlando, R. / Bai, Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2rpi.cif.gz 2rpi.cif.gz | 340.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2rpi.ent.gz pdb2rpi.ent.gz | 279.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2rpi.json.gz 2rpi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rp/2rpi https://data.pdbj.org/pub/pdb/validation_reports/rp/2rpi ftp://data.pdbj.org/pub/pdb/validation_reports/rp/2rpi ftp://data.pdbj.org/pub/pdb/validation_reports/rp/2rpi | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  / RNase H / RNase HMass: 12878.809 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)    Thermus thermophilus HB8 (bacteria) / Gene: rnhA, TTHA1556 / Plasmid: pET42b(+) / Production host: Thermus thermophilus HB8 (bacteria) / Gene: rnhA, TTHA1556 / Plasmid: pET42b(+) / Production host:   Escherichia coli (E. coli) / References: UniProt: P29253, Escherichia coli (E. coli) / References: UniProt: P29253,  ribonuclease H ribonuclease H |
|---|---|
| Sequence details | THE RESIDUES 25-45 IN THE DATABASE SEQUENCE (RNH_THET8 P29253) WERE DELETED SINCE THIS IS THE ...THE RESIDUES 25-45 IN THE DATABASE SEQUENCE (RNH_THET8 P29253) WERE DELETED SINCE THIS IS THE INTERMEDIA |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||
| Sample conditions | Ionic strength: 0.1 / pH: 5.2 / Pressure: ambient / Temperature: 295 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DGSA-distance geometry simulated annealing, torsion angle dynamics Software ordinal: 1 | ||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 10 / Representative conformer: 1 |
 Movie
Movie Controller
Controller












 PDBj
PDBj
