[English] 日本語
 Yorodumi
Yorodumi- PDB-2r9s: c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2r9s | ||||||
|---|---|---|---|---|---|---|---|
| Title | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | ||||||
 Components Components | Mitogen-activated protein kinase 10 | ||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  TRANSFERASE / TRANSFERASE /  jnk3 jnk3 | ||||||
| Function / homology |  Function and homology information Function and homology informationJUN kinase activity / Activation of the AP-1 family of transcription factors / Fc-epsilon receptor signaling pathway /  MAP kinase kinase activity / response to light stimulus / MAP kinase kinase activity / response to light stimulus /  mitogen-activated protein kinase / JNK cascade / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / FCERI mediated MAPK activation / mitogen-activated protein kinase / JNK cascade / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / FCERI mediated MAPK activation /  regulation of circadian rhythm ...JUN kinase activity / Activation of the AP-1 family of transcription factors / Fc-epsilon receptor signaling pathway / regulation of circadian rhythm ...JUN kinase activity / Activation of the AP-1 family of transcription factors / Fc-epsilon receptor signaling pathway /  MAP kinase kinase activity / response to light stimulus / MAP kinase kinase activity / response to light stimulus /  mitogen-activated protein kinase / JNK cascade / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / FCERI mediated MAPK activation / mitogen-activated protein kinase / JNK cascade / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / FCERI mediated MAPK activation /  regulation of circadian rhythm / regulation of circadian rhythm /  cellular senescence / rhythmic process / Oxidative Stress Induced Senescence / cellular senescence / rhythmic process / Oxidative Stress Induced Senescence /  protein phosphorylation / protein serine kinase activity / protein phosphorylation / protein serine kinase activity /  signal transduction / signal transduction /  mitochondrion / mitochondrion /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.4 Å SYNCHROTRON / Resolution: 2.4 Å | ||||||
 Authors Authors | Habel, J. | ||||||
 Citation Citation |  Journal: Bioorg.Med.Chem.Lett. / Year: 2007 Journal: Bioorg.Med.Chem.Lett. / Year: 2007Title: 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors. Authors: Jiang, R. / Duckett, D. / Chen, W. / Habel, J. / Ling, Y.Y. / LoGrasso, P. / Kamenecka, T.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2r9s.cif.gz 2r9s.cif.gz | 157.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2r9s.ent.gz pdb2r9s.ent.gz | 122.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2r9s.json.gz 2r9s.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r9/2r9s https://data.pdbj.org/pub/pdb/validation_reports/r9/2r9s ftp://data.pdbj.org/pub/pdb/validation_reports/r9/2r9s ftp://data.pdbj.org/pub/pdb/validation_reports/r9/2r9s | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1jnkS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||
| 2 | 
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: ASN / Beg label comp-ID: ASN / End auth comp-ID: SER / End label comp-ID: SER / Refine code: 4 / Auth seq-ID: 46 - 401 / Label seq-ID: 1 - 356
| ||||||||||||||||||
| Details | The biological unit is a monomer. There are two biological units in the assymetric unit (chains A & B). |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 41284.789 Da / Num. of mol.: 2 / Fragment: residues 39-402 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: MAPK10, JNK3, JNK3A, PRKM10 / Plasmid: pdest14 / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Gene: MAPK10, JNK3, JNK3A, PRKM10 / Plasmid: pdest14 / Species (production host): Escherichia coli / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3)References: UniProt: P53779,  mitogen-activated protein kinase mitogen-activated protein kinase |
|---|
-Non-polymers , 5 types, 219 molecules 

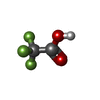






| #2: Chemical | | #3: Chemical | ChemComp-UNX / #4: Chemical |  Trifluoroacetic acid Trifluoroacetic acid#5: Chemical | ChemComp-EDO /  Ethylene glycol Ethylene glycol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.08 Å3/Da / Density % sol: 40.9 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: microbatch / pH: 5.5 Details: 25-32% PEG 3350, 100mM NaCl, 1mM AMP-PCP, 2mM MgCl2, 0.4mM Zwittergent 314, 10%(v/v) ethylene glycol, pH 5.5, microbatch, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL11-1 / Wavelength: 0.997 Å / Beamline: BL11-1 / Wavelength: 0.997 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Apr 10, 2007 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Silicon (111) / Protocol: SINGLE WAVELENGTH / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.997 Å / Relative weight: 1 : 0.997 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.27→50 Å / Num. obs: 32372 / % possible obs: 99.2 % / Redundancy: 6.9 % / Biso Wilson estimate: 41.67 Å2 / Rmerge(I) obs: 0.056 / Χ2: 1.174 / Net I/σ(I): 15.6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Starting model: PDB Entry 1JNK Resolution: 2.4→38.63 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.905 / SU B: 18.952 / SU ML: 0.23 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.624 / ESU R Free: 0.329 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.866 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→38.63 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | Dom-ID: 1 / Auth asym-ID: A / Ens-ID: 1 / Number: 2681 / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→2.462 Å / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj






