[English] 日本語
 Yorodumi
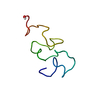
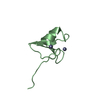
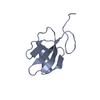
Yorodumi- PDB-2jq8: Solution structure of the Somatomedin B domain from vitronectin p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2jq8 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of the Somatomedin B domain from vitronectin produced in Pichia pastoris | ||||||
 Components Components | Vitronectin | ||||||
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  somatomedin B domain / somatomedin B domain /  vitronectin / disulfide-rich domain / vitronectin / disulfide-rich domain /  PAI-1 / PAI-1 /  uPAR uPAR | ||||||
| Function / homology |  Function and homology information Function and homology informationrough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion /  peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation / peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation /  extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway ...rough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion / extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway ...rough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion /  peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation / peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation /  extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway / positive regulation of cell-substrate adhesion / Molecules associated with elastic fibres / cell adhesion mediated by integrin / extracellular matrix structural constituent / Syndecan interactions / extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway / positive regulation of cell-substrate adhesion / Molecules associated with elastic fibres / cell adhesion mediated by integrin / extracellular matrix structural constituent / Syndecan interactions /  polysaccharide binding / positive regulation of wound healing / positive regulation of smooth muscle cell migration / oligodendrocyte differentiation / endodermal cell differentiation / protein polymerization / polysaccharide binding / positive regulation of wound healing / positive regulation of smooth muscle cell migration / oligodendrocyte differentiation / endodermal cell differentiation / protein polymerization /  basement membrane / ECM proteoglycans / Integrin cell surface interactions / negative regulation of fibrinolysis / basement membrane / ECM proteoglycans / Integrin cell surface interactions / negative regulation of fibrinolysis /  regulation of cell adhesion / regulation of cell adhesion /  collagen binding / extracellular matrix organization / cell-matrix adhesion / collagen binding / extracellular matrix organization / cell-matrix adhesion /  liver regeneration / liver regeneration /  Regulation of Complement cascade / Golgi lumen / positive regulation of receptor-mediated endocytosis / Regulation of Complement cascade / Golgi lumen / positive regulation of receptor-mediated endocytosis /  cell migration / positive regulation of peptidyl-tyrosine phosphorylation / cell migration / positive regulation of peptidyl-tyrosine phosphorylation /  integrin binding / integrin binding /  heparin binding / positive regulation of protein binding / collagen-containing extracellular matrix / blood microparticle / heparin binding / positive regulation of protein binding / collagen-containing extracellular matrix / blood microparticle /  cell adhesion / cell adhesion /  immune response / intracellular membrane-bounded organelle / immune response / intracellular membrane-bounded organelle /  endoplasmic reticulum / endoplasmic reticulum /  extracellular space / extracellular exosome / extracellular region / identical protein binding extracellular space / extracellular exosome / extracellular region / identical protein bindingSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  molecular dynamics molecular dynamics | ||||||
 Authors Authors | Gaardsvoll, H. / Hirschberg, D. / Nielbo, S. / Mayasundari, A. / Peterson, C.B. / Jansson, A. / Jorgensen, T.J.D. / Poulsen, F.M. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2007 Journal: Protein Sci. / Year: 2007Title: Solution structure of recombinant somatomedin B domain from vitronectin produced in Pichia pastoris Authors: Kjaergaard, M. / Gardsvoll, H. / Hirschberg, D. / Nielbo, S. / Mayasundari, A. / Peterson, C.B. / Jansson, A. / Jorgensen, T.J.D. / Poulsen, F.M. / Ploug, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2jq8.cif.gz 2jq8.cif.gz | 271.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2jq8.ent.gz pdb2jq8.ent.gz | 224.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2jq8.json.gz 2jq8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jq/2jq8 https://data.pdbj.org/pub/pdb/validation_reports/jq/2jq8 ftp://data.pdbj.org/pub/pdb/validation_reports/jq/2jq8 ftp://data.pdbj.org/pub/pdb/validation_reports/jq/2jq8 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  / Serum-spreading factor / S-protein / V75 / Serum-spreading factor / S-protein / V75Mass: 6174.770 Da / Num. of mol.: 1 / Fragment: sequence database residues 20-66 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: VTN / Production host: Homo sapiens (human) / Gene: VTN / Production host:   Pichia pastoris (fungus) / Strain (production host): X-33 / References: UniProt: P04004 Pichia pastoris (fungus) / Strain (production host): X-33 / References: UniProt: P04004 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||
| Sample conditions | Ionic strength: 0 / pH: 6.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Varian Unity / Manufacturer: Varian / Model : UNITY / Field strength: 750 MHz : UNITY / Field strength: 750 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  molecular dynamics / Software ordinal: 1 molecular dynamics / Software ordinal: 1 | ||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj






