+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1s4g | ||||||
|---|---|---|---|---|---|---|---|
| Title | Somatomedin-B Domain of human plasma vitronectin. | ||||||
 Components Components | Vitronectin | ||||||
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  Somatomedin B domain / disulfide knot / Somatomedin B domain / disulfide knot /  vitronectin vitronectin | ||||||
| Function / homology |  Function and homology information Function and homology informationrough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion /  peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation / peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation /  extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway ...rough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion / extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway ...rough endoplasmic reticulum lumen / smooth muscle cell-matrix adhesion /  peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation / peptidase inhibitor complex / alphav-beta3 integrin-vitronectin complex / scavenger receptor activity / negative regulation of endopeptidase activity / protein complex involved in cell-matrix adhesion / negative regulation of blood coagulation /  extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway / positive regulation of cell-substrate adhesion / Molecules associated with elastic fibres / cell adhesion mediated by integrin / extracellular matrix structural constituent / Syndecan interactions / extracellular matrix binding / positive regulation of vascular endothelial growth factor receptor signaling pathway / positive regulation of cell-substrate adhesion / Molecules associated with elastic fibres / cell adhesion mediated by integrin / extracellular matrix structural constituent / Syndecan interactions /  polysaccharide binding / positive regulation of wound healing / positive regulation of smooth muscle cell migration / oligodendrocyte differentiation / endodermal cell differentiation / protein polymerization / polysaccharide binding / positive regulation of wound healing / positive regulation of smooth muscle cell migration / oligodendrocyte differentiation / endodermal cell differentiation / protein polymerization /  basement membrane / ECM proteoglycans / Integrin cell surface interactions / negative regulation of fibrinolysis / basement membrane / ECM proteoglycans / Integrin cell surface interactions / negative regulation of fibrinolysis /  regulation of cell adhesion / regulation of cell adhesion /  collagen binding / extracellular matrix organization / cell-matrix adhesion / collagen binding / extracellular matrix organization / cell-matrix adhesion /  liver regeneration / liver regeneration /  Regulation of Complement cascade / Golgi lumen / positive regulation of receptor-mediated endocytosis / Regulation of Complement cascade / Golgi lumen / positive regulation of receptor-mediated endocytosis /  cell migration / positive regulation of peptidyl-tyrosine phosphorylation / cell migration / positive regulation of peptidyl-tyrosine phosphorylation /  integrin binding / integrin binding /  heparin binding / positive regulation of protein binding / collagen-containing extracellular matrix / blood microparticle / heparin binding / positive regulation of protein binding / collagen-containing extracellular matrix / blood microparticle /  cell adhesion / cell adhesion /  immune response / intracellular membrane-bounded organelle / immune response / intracellular membrane-bounded organelle /  endoplasmic reticulum / endoplasmic reticulum /  extracellular space / extracellular exosome / extracellular region / identical protein binding extracellular space / extracellular exosome / extracellular region / identical protein bindingSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
 Authors Authors | Mayasundari, A. / Whittemore, N.A. / Serpersu, E.H. / Peterson, C.B. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2004 Journal: J.Biol.Chem. / Year: 2004Title: The solution structure of the N-terminal domain of human vitronectin: proximal sites that regulate fibrinolysis and cell migration Authors: Mayasundari, A. / Whittemore, N.A. / Serpersu, E.H. / Peterson, C.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1s4g.cif.gz 1s4g.cif.gz | 297 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1s4g.ent.gz pdb1s4g.ent.gz | 255.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1s4g.json.gz 1s4g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/s4/1s4g https://data.pdbj.org/pub/pdb/validation_reports/s4/1s4g ftp://data.pdbj.org/pub/pdb/validation_reports/s4/1s4g ftp://data.pdbj.org/pub/pdb/validation_reports/s4/1s4g | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 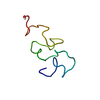
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  Mass: 5824.493 Da / Num. of mol.: 1 / Fragment: Somatomedin B / Source method: isolated from a natural source / Source: (natural)   Homo sapiens (human) / Tissue: plasma / References: UniProt: P04004 Homo sapiens (human) / Tissue: plasma / References: UniProt: P04004 |
|---|---|
| #2: Chemical | ChemComp-OH /  Hydroxide Hydroxide |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: Isolated from human plasma vitronectin by CNBr cleavage |
|---|---|
| Sample conditions | Ionic strength: low / pH: 4.4 / Temperature: 298 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model : INOVA / Field strength: 600 MHz : INOVA / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 Details: NOE cross-peak intensities were converted into distance restraints as follows: strong, 1.8-2.7 ; medium, 1.8-3.4 ; weak, 1.8-4.5 , and very weak 1.8-6.0. An additional 1.0 was added to upper ...Details: NOE cross-peak intensities were converted into distance restraints as follows: strong, 1.8-2.7 ; medium, 1.8-3.4 ; weak, 1.8-4.5 , and very weak 1.8-6.0. An additional 1.0 was added to upper limits involving methyl protons, 0.5 for methylene protons and 2.3 for degenerate Hd and He protons of tyrosines and phenylalanines. Also, a 0.2 was added to the upper limits of NOEs involving amide protons. Backbone F angles were restrained to -120 50 for 3JHNHa = 8-9 Hz, and -120 40 for JHNHa > 9Hz. A restraint of 100 80 was also applied to F angle for residues that show stronger NHi-Hai-1 NOE than the intraresidue NH-Ha NOE. A total of 329 NOE restraints and 18 F restraints were used in structure determination. Random structures were generated by subjecting the peptide to an initial 10000-step minimization at 298K. The temperature was then raised gradually to 1000K during a 1000 step dynamics simulation. The peptide was subjected to minimization and a 10ps dynamics at 1000K. The NMR-derived restraints were then imposed on the peptide and the peptide was slowly annealed to 298K in a 100ps trajectory. Finally, the structures were subjected to further minimization at 298K. The force constant for the distance restraints was 100 kcal/mol 2 and the dielectric constant was 4. | ||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with favorable non-bond energy Conformers calculated total number: 60 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller










 PDBj
PDBj