[English] 日本語
 Yorodumi
Yorodumi- PDB-2j7q: Crystal structure of the ubiquitin-specific protease encoded by m... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2j7q | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
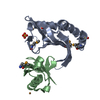
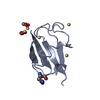
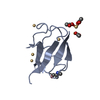
| Title | Crystal structure of the ubiquitin-specific protease encoded by murine cytomegalovirus tegument protein M48 in complex with a ubquitin-based suicide substrate | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  HERPESVIRIDAE / HERPESVIRIDAE /  NUCLEAR PROTEIN / COVALENT ENZYME-LIGAND COMPLEX / NUCLEAR PROTEIN / COVALENT ENZYME-LIGAND COMPLEX /  DEUBIQUITINATING ENZYME / PAPAIN-LIKE FOLD / DEUBIQUITINATING ENZYME / PAPAIN-LIKE FOLD /  CYSTEINE PROTEASE CYSTEINE PROTEASE | |||||||||
| Function / homology |  Function and homology information Function and homology information : / : /  : / protein modification process => GO:0036211 / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation ... : / protein modification process => GO:0036211 / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation ... : / : /  : / protein modification process => GO:0036211 / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Prevention of phagosomal-lysosomal fusion / IRAK2 mediated activation of TAK1 complex / Alpha-protein kinase 1 signaling pathway / : / protein modification process => GO:0036211 / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Prevention of phagosomal-lysosomal fusion / IRAK2 mediated activation of TAK1 complex / Alpha-protein kinase 1 signaling pathway /  Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Membrane binding and targetting of GAG proteins / Endosomal Sorting Complex Required For Transport (ESCRT) / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Negative regulation of FLT3 / Constitutive Signaling by NOTCH1 HD Domain Mutants / Regulation of FZD by ubiquitination / TICAM1,TRAF6-dependent induction of TAK1 complex / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / p75NTR recruits signalling complexes / Downregulation of ERBB4 signaling / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / APC-Cdc20 mediated degradation of Nek2A / PINK1-PRKN Mediated Mitophagy / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Pexophagy / Regulation of innate immune responses to cytosolic DNA / InlA-mediated entry of Listeria monocytogenes into host cells / VLDLR internalisation and degradation / cytosolic ribosome / Downregulation of ERBB2:ERBB3 signaling / NF-kB is activated and signals survival / NRIF signals cell death from the nucleus / Regulation of PTEN localization / Activated NOTCH1 Transmits Signal to the Nucleus / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Membrane binding and targetting of GAG proteins / Endosomal Sorting Complex Required For Transport (ESCRT) / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Negative regulation of FLT3 / Constitutive Signaling by NOTCH1 HD Domain Mutants / Regulation of FZD by ubiquitination / TICAM1,TRAF6-dependent induction of TAK1 complex / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1-dependent activation of IRF3/IRF7 / APC/C:Cdc20 mediated degradation of Cyclin B / p75NTR recruits signalling complexes / Downregulation of ERBB4 signaling / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / APC-Cdc20 mediated degradation of Nek2A / PINK1-PRKN Mediated Mitophagy / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Pexophagy / Regulation of innate immune responses to cytosolic DNA / InlA-mediated entry of Listeria monocytogenes into host cells / VLDLR internalisation and degradation / cytosolic ribosome / Downregulation of ERBB2:ERBB3 signaling / NF-kB is activated and signals survival / NRIF signals cell death from the nucleus / Regulation of PTEN localization / Activated NOTCH1 Transmits Signal to the Nucleus /  Regulation of BACH1 activity / Translesion synthesis by REV1 / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Translesion synthesis by POLK / MAP3K8 (TPL2)-dependent MAPK1/3 activation / TICAM1, RIP1-mediated IKK complex recruitment / Downregulation of TGF-beta receptor signaling / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / Josephin domain DUBs / Regulation of activated PAK-2p34 by proteasome mediated degradation / InlB-mediated entry of Listeria monocytogenes into host cell / IKK complex recruitment mediated by RIP1 / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Autodegradation of Cdh1 by Cdh1:APC/C / TNFR1-induced NF-kappa-B signaling pathway / APC/C:Cdc20 mediated degradation of Securin / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / TCF dependent signaling in response to WNT / NIK-->noncanonical NF-kB signaling / Regulation of NF-kappa B signaling / Ubiquitin-dependent degradation of Cyclin D / AUF1 (hnRNP D0) binds and destabilizes mRNA / TNFR2 non-canonical NF-kB pathway / activated TAK1 mediates p38 MAPK activation / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / NOTCH3 Activation and Transmission of Signal to the Nucleus / Negative regulators of DDX58/IFIH1 signaling / Deactivation of the beta-catenin transactivating complex / Degradation of DVL / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Regulation of signaling by CBL / Dectin-1 mediated noncanonical NF-kB signaling / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Fanconi Anemia Pathway / Hh mutants are degraded by ERAD / Negative regulation of FGFR3 signaling / Recognition of DNA damage by PCNA-containing replication complex / Regulation of BACH1 activity / Translesion synthesis by REV1 / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Translesion synthesis by POLK / MAP3K8 (TPL2)-dependent MAPK1/3 activation / TICAM1, RIP1-mediated IKK complex recruitment / Downregulation of TGF-beta receptor signaling / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Translesion synthesis by POLI / Gap-filling DNA repair synthesis and ligation in GG-NER / Josephin domain DUBs / Regulation of activated PAK-2p34 by proteasome mediated degradation / InlB-mediated entry of Listeria monocytogenes into host cell / IKK complex recruitment mediated by RIP1 / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Autodegradation of Cdh1 by Cdh1:APC/C / TNFR1-induced NF-kappa-B signaling pathway / APC/C:Cdc20 mediated degradation of Securin / Asymmetric localization of PCP proteins / SCF-beta-TrCP mediated degradation of Emi1 / TCF dependent signaling in response to WNT / NIK-->noncanonical NF-kB signaling / Regulation of NF-kappa B signaling / Ubiquitin-dependent degradation of Cyclin D / AUF1 (hnRNP D0) binds and destabilizes mRNA / TNFR2 non-canonical NF-kB pathway / activated TAK1 mediates p38 MAPK activation / Assembly of the pre-replicative complex / Vpu mediated degradation of CD4 / NOTCH3 Activation and Transmission of Signal to the Nucleus / Negative regulators of DDX58/IFIH1 signaling / Deactivation of the beta-catenin transactivating complex / Degradation of DVL / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Regulation of signaling by CBL / Dectin-1 mediated noncanonical NF-kB signaling / Cdc20:Phospho-APC/C mediated degradation of Cyclin A / Fanconi Anemia Pathway / Hh mutants are degraded by ERAD / Negative regulation of FGFR3 signaling / Recognition of DNA damage by PCNA-containing replication complex /  Peroxisomal protein import / Degradation of AXIN / Downregulation of SMAD2/3:SMAD4 transcriptional activity / Degradation of GLI1 by the proteasome Peroxisomal protein import / Degradation of AXIN / Downregulation of SMAD2/3:SMAD4 transcriptional activity / Degradation of GLI1 by the proteasomeSimilarity search - Function | |||||||||
| Biological species |   MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus) MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus)  HOMO SAPIENS (human) HOMO SAPIENS (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 1.8 Å SAD / Resolution: 1.8 Å | |||||||||
 Authors Authors | Schlieker, C. / Weihofen, W.A. / Frijns, E. / Kattenhorn, L.M. / Gaudet, R. / Ploegh, H.L. | |||||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2007 Journal: Mol.Cell / Year: 2007Title: Structure of a Herpesvirus-Encoded Cysteine Protease Reveals a Unique Class of Deubiquitinating Enzymes Authors: Schlieker, C. / Weihofen, W.A. / Frijns, E. / Kattenhorn, L.M. / Gaudet, R. / Ploegh, H.L. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN ... SHEET THE SHEET STRUCTURE OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS ARE DEFINED. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2j7q.cif.gz 2j7q.cif.gz | 152.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2j7q.ent.gz pdb2j7q.ent.gz | 118.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2j7q.json.gz 2j7q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j7/2j7q https://data.pdbj.org/pub/pdb/validation_reports/j7/2j7q ftp://data.pdbj.org/pub/pdb/validation_reports/j7/2j7q ftp://data.pdbj.org/pub/pdb/validation_reports/j7/2j7q | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Noncrystallographic symmetry (NCS) | NCS oper:
|
- Components
Components
-MCMV TEGUMENT PROTEIN M48 ENCODED UBIQUITIN- SPECIFIC PROTEASE, ... , 2 types, 2 molecules AC
| #1: Protein | Mass: 25677.289 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: DEUBIQUITINATING MODULE OF MURINE CYTOMEGALOVIRUS TEGUMENT PROTEIN M48. ACTIVE SITE CYSTEINE 23 IS COVALENTLY LINKED TO THE FORMER VINYLMETHYLESTER MOIETY OF THE SUICIDE SUBSTATE UBVME Source: (gene. exp.)   MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus) MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus)Strain: MCMV STRAIN SMITH / Plasmid: PET28 (NOVAGEN) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) |
|---|---|
| #3: Protein | Mass: 25686.316 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: DEUBIQUITINATING MODULE OF MURINE CYTOMEGALOVIRUS TEGUMENT PROTEIN M48. ACTIVE SITE CYSTEINE 23 IS COVALENTLY LINKED TO THE FORMER VINYLMETHYLESTER MOIETY OF THE SUICIDE SUBSTATE UBVME Source: (gene. exp.)   MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus) MURINE CYTOMEGALOVIRUS (Murine cytomegalovirus)Strain: MCMV STRAIN SMITH / Plasmid: PET28 (NOVAGEN) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) |
-Protein , 1 types, 2 molecules BD
| #2: Protein |  Mass: 8519.778 Da / Num. of mol.: 2 Fragment: UBIQUITIN FUSED TO VINYLMETHYLESTER, UBVME, RESIDUES 1-75 Source method: isolated from a genetically manipulated source Details: THE C-TERMINAL GLY 76 IS REPLACED BY THE VINYLMETHYLESTER MOIETY Source: (gene. exp.)   HOMO SAPIENS (human) / Plasmid: PTYB (NEW ENGLAND BIOLABS) / Production host: HOMO SAPIENS (human) / Plasmid: PTYB (NEW ENGLAND BIOLABS) / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P62988, UniProt: P0CG48*PLUS ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P62988, UniProt: P0CG48*PLUS |
|---|
-Non-polymers , 5 types, 714 molecules 








| #4: Chemical | | #5: Chemical | #6: Chemical | ChemComp-PG4 / |  Polyethylene glycol Polyethylene glycol#7: Chemical | ChemComp-GOL / |  Glycerol Glycerol#8: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.95 Å3/Da / Density % sol: 36.49 % |
|---|---|
Crystal grow | pH: 7.5 / Details: 200 MM MAGNESIUM FORMATE, 14% PEG 3350, pH 7.50 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.9797 / Beamline: 24-ID-C / Wavelength: 0.9797 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Mar 31, 2006 |
| Radiation | Monochromator: SI 111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9797 Å / Relative weight: 1 : 0.9797 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→50 Å / Num. obs: 52323 / % possible obs: 97.4 % / Observed criterion σ(I): 2 / Redundancy: 4.2 % / Rmerge(I) obs: 0.09 / Net I/σ(I): 8.3 |
| Reflection shell | Resolution: 1.8→1.86 Å / Redundancy: 2.4 % / Rmerge(I) obs: 0.27 / Mean I/σ(I) obs: 3.5 / % possible all: 84.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 1.8→50 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.925 / SU B: 4.84 / SU ML: 0.079 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.123 / ESU R Free: 0.128 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. SAD / Resolution: 1.8→50 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.925 / SU B: 4.84 / SU ML: 0.079 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.123 / ESU R Free: 0.128 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.21 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→50 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






































 PDBj
PDBj


























