+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tdg | ||||||
|---|---|---|---|---|---|---|---|
| Title | Complex of S130G SHV-1 beta-lactamase with tazobactam | ||||||
 Components Components | Beta-lactamase SHV-1 | ||||||
 Keywords Keywords |  HYDROLASE / S130G SHV-1 class A beta-lactamase / HYDROLASE / S130G SHV-1 class A beta-lactamase /  penicillinase / penicillinase /  beta-lactam hydrolase / beta-lactam hydrolase /  detergent binding / detergent binding /  inhibitor complex / inhibitor complex /  tazobactam / tazobactam /  aldehyde / cymal-6 aldehyde / cymal-6 | ||||||
| Function / homology |  Function and homology information Function and homology informationbeta-lactam antibiotic catabolic process /  beta-lactamase activity / beta-lactamase activity /  beta-lactamase / response to antibiotic beta-lactamase / response to antibioticSimilarity search - Function | ||||||
| Biological species |   Klebsiella pneumoniae (bacteria) Klebsiella pneumoniae (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Sun, T. / Bethel, C.R. / Bonomo, R.A. / Knox, J.R. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2004 Journal: Biochemistry / Year: 2004Title: Inhibitor-resistant class A beta-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant Authors: Sun, T. / Bethel, C.R. / Bonomo, R.A. / Knox, J.R. | ||||||
| History |
| ||||||
| Remark 600 | HETEROGEN Apparent short contacts between the 2 intermediates, TBE and MDD, arise because the ...HETEROGEN Apparent short contacts between the 2 intermediates, TBE and MDD, arise because the crystal structure contains a composite of 2 complexes, one a breakdown product of the other. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tdg.cif.gz 1tdg.cif.gz | 82.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tdg.ent.gz pdb1tdg.ent.gz | 59.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tdg.json.gz 1tdg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/td/1tdg https://data.pdbj.org/pub/pdb/validation_reports/td/1tdg ftp://data.pdbj.org/pub/pdb/validation_reports/td/1tdg ftp://data.pdbj.org/pub/pdb/validation_reports/td/1tdg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1tdlC  1shvS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 28876.994 Da / Num. of mol.: 1 / Mutation: S130G Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Klebsiella pneumoniae (bacteria) / Gene: bla / Plasmid: pBC SK / Species (production host): Escherichia coli Klebsiella pneumoniae (bacteria) / Gene: bla / Plasmid: pBC SK / Species (production host): Escherichia coliProduction host:   Escherichia coli str. K-12 substr. DH10B (bacteria) Escherichia coli str. K-12 substr. DH10B (bacteria)Strain (production host): DH10B References: UniProt: P14557, UniProt: P0AD64*PLUS,  beta-lactamase beta-lactamase |
|---|
-Non-polymers , 6 types, 402 molecules 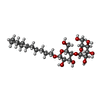










| #2: Chemical | | #3: Chemical | ChemComp-TBE / | #4: Chemical | ChemComp-MDD / |  Malondialdehyde Malondialdehyde#5: Chemical | ChemComp-MRD / ( |  2-Methyl-2,4-pentanediol 2-Methyl-2,4-pentanediol#6: Chemical | ChemComp-MPD / ( |  2-Methyl-2,4-pentanediol 2-Methyl-2,4-pentanediol#7: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.99 Å3/Da / Density % sol: 38 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 15% PEG 6000, 50 mM HEPES, 0.56 mM Cymal-6 detergent, pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å |
| Detector | Type: SIEMENS HI-STAR / Detector: AREA DETECTOR / Date: Jul 1, 2003 / Details: double-mirror Franks focusing |
| Radiation | Monochromator: Ni FILTER / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→28 Å / Num. all: 22043 / Num. obs: 20318 / % possible obs: 92.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 6.1 % / Biso Wilson estimate: 15.7 Å2 / Rmerge(I) obs: 0.059 / Rsym value: 0.059 / Net I/σ(I): 14.8 |
| Reflection shell | Resolution: 1.8→1.91 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.138 / Mean I/σ(I) obs: 4.5 / Num. unique all: 3624 / Rsym value: 0.138 / % possible all: 79.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1SHV Resolution: 1.8→20 Å / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber Details: Atoms having alternate conformations are only listed once in the above list of the number of atoms used in the refinement
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 10.9 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→20 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj







