[English] 日本語
 Yorodumi
Yorodumi- PDB-1c5f: CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c5f | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALAYI COMPLEXED WITH CYCLOSPORIN A | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | ISOMERASE/IMMUNOSUPPRESSANT / ISOMERASE-IMMUNOSUPPRESSANT COMPLEX / CYCLOPHILIN-CYCLOSPORIN COMPLEX /  CYCLOSPORIN A / CYCLOSPORIN A /  IMMUNOSUPPRESSANT / IMMUNOSUPPRESSANT /  CYCLOPHILIN CYCLOPHILIN | |||||||||
| Function / homology |  Function and homology information Function and homology information peptidylprolyl isomerase / peptidylprolyl isomerase /  peptidyl-prolyl cis-trans isomerase activity / peptidyl-prolyl cis-trans isomerase activity /  protein folding / intracellular membrane-bounded organelle / protein folding / intracellular membrane-bounded organelle /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   BRUGIA MALAYI (agent of lymphatic filariasis) BRUGIA MALAYI (agent of lymphatic filariasis)  TOLYPOCLADIUM INFLATUM (fungus) TOLYPOCLADIUM INFLATUM (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.47 Å MOLECULAR REPLACEMENT / Resolution: 2.47 Å | |||||||||
 Authors Authors | Ellis, P.J. / Carlow, C.K.S. / Ma, D. / Kuhn, P. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2000 Journal: Biochemistry / Year: 2000Title: Crystal Structure of the Complex of Brugia Malayi Cyclophilin and Cyclosporin A. Authors: Ellis, P.J. / Carlow, C.K. / Ma, D. / Kuhn, P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c5f.cif.gz 1c5f.cif.gz | 293.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c5f.ent.gz pdb1c5f.ent.gz | 244.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c5f.json.gz 1c5f.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c5/1c5f https://data.pdbj.org/pub/pdb/validation_reports/c5/1c5f ftp://data.pdbj.org/pub/pdb/validation_reports/c5/1c5f ftp://data.pdbj.org/pub/pdb/validation_reports/c5/1c5f | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 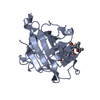
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| 7 | 
| ||||||||
| 8 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Prolyl isomerase / E.C.5.2.1.8 / PPIASE 1 / ROTAMASE / CYCLOPHILIN Prolyl isomerase / E.C.5.2.1.8 / PPIASE 1 / ROTAMASE / CYCLOPHILINMass: 19504.418 Da / Num. of mol.: 8 / Fragment: CYCLOPHILIN-LIKE DOMAIN, RESIDUES 1-177 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   BRUGIA MALAYI (agent of lymphatic filariasis) BRUGIA MALAYI (agent of lymphatic filariasis)Gene: BMCYP-1 / Plasmid: PMAL-C2 / Gene (production host): MBP FUSION PROTEIN / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): ER2267 / References: UniProt: Q27450, ESCHERICHIA COLI (E. coli) / Strain (production host): ER2267 / References: UniProt: Q27450,  peptidylprolyl isomerase peptidylprolyl isomerase#2: Protein/peptide |  Ciclosporin / CYCLOSPORINE / CICLOSPORIN / CICLOSPORINE / Ciclosporin / CYCLOSPORINE / CICLOSPORIN / CICLOSPORINE /  Ciclosporin Ciclosporin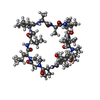  Type: Cyclic peptide Type: Cyclic peptide / Class: Immunosuppressant / Class: Immunosuppressant Immunosuppressive drug / Mass: 1220.625 Da / Num. of mol.: 8 / Source method: obtained synthetically Immunosuppressive drug / Mass: 1220.625 Da / Num. of mol.: 8 / Source method: obtained syntheticallyDetails: CYCLOSPORIN IS A CYCLIC UNDECAPEPTIDE. CYCLIZATION IS ACHIEVED BY LINKING THE N- AND THE C- TERMINI. Source: (synth.)   TOLYPOCLADIUM INFLATUM (fungus) / References: TOLYPOCLADIUM INFLATUM (fungus) / References:  NOR: NOR00033, NOR: NOR00033,  Cyclosporin A Cyclosporin A#3: Water | ChemComp-HOH / |  Water WaterCompound details | CYCLOSPORI | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.5 Å3/Da / Density % sol: 50.83 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | pH: 6 / Details: PEG 8000, CALCIUM ACETATE, METHANOL, MES, PH 6.00 | |||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 22 ℃ / pH: 6 / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 105 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-1 / Wavelength: 0.98 / Beamline: BL9-1 / Wavelength: 0.98 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jul 4, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.98 Å / Relative weight: 1 : 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 2.47→47.14 Å / Num. obs: 56200 / % possible obs: 95.7 % / Observed criterion σ(I): 0 / Redundancy: 2.9 % / Biso Wilson estimate: 53.4 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 9 |
| Reflection shell | Resolution: 2.47→2.53 Å / Redundancy: 1.6 % / Rmerge(I) obs: 0.372 / % possible all: 73.1 |
| Reflection | *PLUS % possible obs: 77.5 % / Observed criterion σ(I): 1 / Redundancy: 2.9 % / Num. measured all: 409247 / Rmerge(I) obs: 0.12 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.47→47.1 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH & HUBER MOLECULAR REPLACEMENT / Resolution: 2.47→47.1 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: ENGH & HUBERDetails: USED MAXIMUM LIKELIHOOD TARGET BASED ON STRUCTURE FACTORS WITH NO CUTOFFS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 31.56 Å2 / ksol: 0.31 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 43.46 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.47→47.1 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.47→2.58 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 47.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller









































 PDBj
PDBj


