[English] 日本語
 Yorodumi
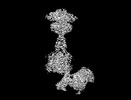
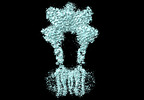
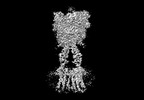
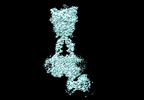
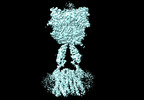
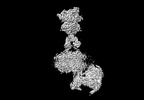
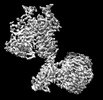
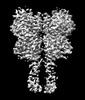
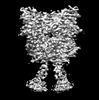
Yorodumi- EMDB-36177: Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex structure / Gi-bound metabotropic glutamate receptor mGlu4 /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationadenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / neurotransmitter secretion /  glutamate receptor activity / GTP metabolic process / dopamine receptor signaling pathway / positive regulation of macroautophagy ...adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / neurotransmitter secretion / glutamate receptor activity / GTP metabolic process / dopamine receptor signaling pathway / positive regulation of macroautophagy ...adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / neurotransmitter secretion /  glutamate receptor activity / GTP metabolic process / dopamine receptor signaling pathway / positive regulation of macroautophagy / Adenylate cyclase inhibitory pathway / regulation of neuron apoptotic process / glutamate receptor activity / GTP metabolic process / dopamine receptor signaling pathway / positive regulation of macroautophagy / Adenylate cyclase inhibitory pathway / regulation of neuron apoptotic process /  regulation of synaptic transmission, glutamatergic / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor activity / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / adenylate cyclase-activating dopamine receptor signaling pathway / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / regulation of synaptic transmission, glutamatergic / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor activity / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / adenylate cyclase-activating dopamine receptor signaling pathway / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade /  heterotrimeric G-protein complex / G alpha (12/13) signalling events / heterotrimeric G-protein complex / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / presynapse / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / presynapse /  GTPase binding / phospholipase C-activating G protein-coupled receptor signaling pathway / Ca2+ pathway / midbody / G alpha (i) signalling events / fibroblast proliferation / cytoplasmic vesicle / G alpha (s) signalling events / G alpha (q) signalling events / chemical synaptic transmission / cell population proliferation / Ras protein signal transduction / positive regulation of MAPK cascade / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / GTPase binding / phospholipase C-activating G protein-coupled receptor signaling pathway / Ca2+ pathway / midbody / G alpha (i) signalling events / fibroblast proliferation / cytoplasmic vesicle / G alpha (s) signalling events / G alpha (q) signalling events / chemical synaptic transmission / cell population proliferation / Ras protein signal transduction / positive regulation of MAPK cascade / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway /  cell cycle / lysosomal membrane / cell cycle / lysosomal membrane /  cell division / cell division /  GTPase activity / GTPase activity /  centrosome / centrosome /  synapse / protein-containing complex binding / synapse / protein-containing complex binding /  nucleolus / GTP binding / nucleolus / GTP binding /  Golgi apparatus / Golgi apparatus /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  nucleoplasm / nucleoplasm /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wang X / Wang M / Xu T / Feng Y / Han S / Lin S / Zhao Q / Wu B | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers. Authors: Xinwei Wang / Mu Wang / Tuo Xu / Ye Feng / Qiang Shao / Shuo Han / Xiaojing Chu / Yechun Xu / Shuling Lin / Qiang Zhao / Beili Wu /  Abstract: Heterodimerization of the metabotropic glutamate receptors (mGlus) has shown importance in the functional modulation of the receptors and offers potential drug targets for treating central nervous ...Heterodimerization of the metabotropic glutamate receptors (mGlus) has shown importance in the functional modulation of the receptors and offers potential drug targets for treating central nervous system diseases. However, due to a lack of molecular details of the mGlu heterodimers, understanding of the mechanisms underlying mGlu heterodimerization and activation is limited. Here we report twelve cryo-electron microscopy (cryo-EM) structures of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers in different conformational states, including inactive, intermediate inactive, intermediate active and fully active conformations. These structures provide a full picture of conformational rearrangement of mGlu2-mGlu3 upon activation. The Venus flytrap domains undergo a sequential conformational change, while the transmembrane domains exhibit a substantial rearrangement from an inactive, symmetric dimer with diverse dimerization patterns to an active, asymmetric dimer in a conserved dimerization mode. Combined with functional data, these structures reveal that stability of the inactive conformations of the subunits and the subunit-G protein interaction pattern are determinants of asymmetric signal transduction of the heterodimers. Furthermore, a novel binding site for two mGlu4 positive allosteric modulators was observed in the asymmetric dimer interfaces of the mGlu2-mGlu4 heterodimer and mGlu4 homodimer, and may serve as a drug recognition site. These findings greatly extend our knowledge about signal transduction of the mGlus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36177.map.gz emd_36177.map.gz | 160.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36177-v30.xml emd-36177-v30.xml emd-36177.xml emd-36177.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36177.png emd_36177.png | 46.2 KB | ||
| Filedesc metadata |  emd-36177.cif.gz emd-36177.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36177 http://ftp.pdbj.org/pub/emdb/structures/EMD-36177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36177 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36177 | HTTPS FTP |
-Related structure data
| Related structure data |  8jd6MC  8jcuC  8jcvC  8jcwC  8jcxC  8jcyC  8jczC  8jd0C  8jd1C  8jd2C  8jd3C  8jd4C  8jd5C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36177.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36177.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Gi1-bound metabotropic glutamate receptor mGlu4
| Entire | Name: Gi1-bound metabotropic glutamate receptor mGlu4 |
|---|---|
| Components |
|
-Supramolecule #1: Gi1-bound metabotropic glutamate receptor mGlu4
| Supramolecule | Name: Gi1-bound metabotropic glutamate receptor mGlu4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Metabotropic glutamate receptor 4
| Macromolecule | Name: Metabotropic glutamate receptor 4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.458359 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: DYKDDDDGAP KPKGHPHMNS IRIDGDITLG GLFPVHGRGS EGKPCGELKK EKGIHRLEAM LFALDRINND PDLLPNITLG ARILDTCSR DTHALEQSLT FVQALIEKDG TEVRCGSGGP PIITKPERVV GVIGASGSSV SIMVANILRL FKIPQISYAS T APDLSDNS ...String: DYKDDDDGAP KPKGHPHMNS IRIDGDITLG GLFPVHGRGS EGKPCGELKK EKGIHRLEAM LFALDRINND PDLLPNITLG ARILDTCSR DTHALEQSLT FVQALIEKDG TEVRCGSGGP PIITKPERVV GVIGASGSSV SIMVANILRL FKIPQISYAS T APDLSDNS RYDFFSRVVP SDTYQAQAMV DIVRALKWNY VSTVASEGSY GESGVEAFIQ KSREDGGVCI AQSVKIPREP KA GEFDKII RRLLETSNAR AVIIFANEDD IRRVLEAARR ANQTGHFFWM GSDSWGSKIA PVLHLEEVAE GAVTILPKRM SVR GFDRYF SSRTLDNNRR NIWFAEFWED NFHCKLSRHA LKKGSHVKKC TNRERIGQDS AYEQEGKVQF VIDAVYAMGH ALHA MHRDL CPGRVGLCPR MDPVDGTQLL KYIRNVNFSG IAGNPVTFNE NGDAPGRYDI YQYQLRNDSA EYKVIGSWTD HLHLR IERM HWPGSGQQLP RSICSLPCQP GERKKTVKGM PCCWHCEPCT GYQYQVDRYT CKTCPYDMRP TENRTGCRPI PIIKLE WGS PWAVLPLFLA VVGIAATLFV VITFVRYNDT PIVKASGREL SYVLLAGIFL CYATTFLMIA EPDLGTCSLR RIFLGLG MS ISYAALLTKT NRIYRIFEQG KRSVSAPRFI SPASQLAITF SLISLQLLGI CVWFVVDPSH SVVDFQDQRT LDPRFARG V LKCDISDLSL ICLLGYSMLL MVTCTVYAIK TRGVPETFNE AKPIGFTMYT TCIVWLAFIP IFFGTSQSAD KLYIQTTTL TVSVSLSASV SLGMLYMPKV YIILFHPEQN VPKRKRSLKA VVTAATMSNK FTQKGNFRPN GEAKSELCEN LEAPALATKQ TYVTYTNHA I UniProtKB:  Metabotropic glutamate receptor 4 Metabotropic glutamate receptor 4 |
-Macromolecule #2: scFv
| Macromolecule | Name: scFv / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 27.409588 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(i) subunit alpha-3
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-3 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.55207 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AKEVKLLLLG AGESGKNTIV KQMKIIHEDG YSEDECKQYK VVVYSNTIQS IIAIIRAMG RLKIDFGEAA RADDARQLFV LAGSAEEGVM TPELAGVIKR LWRDGGVQAC FSRSREYQLN DSASYYLNDL D RISQSNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AKEVKLLLLG AGESGKNTIV KQMKIIHEDG YSEDECKQYK VVVYSNTIQS IIAIIRAMG RLKIDFGEAA RADDARQLFV LAGSAEEGVM TPELAGVIKR LWRDGGVQAC FSRSREYQLN DSASYYLNDL D RISQSNYI PTQQDVLRTR VKTTGIVETH FTDKDLYFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTETSIILF LNKKDLFEEK IKRSPLTICY PEYTGSNTYE EAAAYIQCQF EDLNRRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKECGLY UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-3 |
-Macromolecule #6: N-(3-chlorophenyl)pyridine-2-carboxamide
| Macromolecule | Name: N-(3-chlorophenyl)pyridine-2-carboxamide / type: ligand / ID: 6 / Number of copies: 1 / Formula: BK0 |
|---|---|
| Molecular weight | Theoretical: 232.666 Da |
| Chemical component information |  ChemComp-BK0: |
-Macromolecule #7: PHOSPHOSERINE
| Macromolecule | Name: PHOSPHOSERINE / type: ligand / ID: 7 / Number of copies: 2 / Formula: SEP |
|---|---|
| Molecular weight | Theoretical: 185.072 Da |
| Chemical component information | 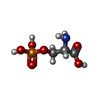 ChemComp-SEP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 839200 |
 Movie
Movie Controller
Controller