[English] 日本語
 Yorodumi
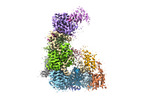
Yorodumi- EMDB-31742: Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex -
+ Open data
Open data
- Basic information
Basic information
| Entry | 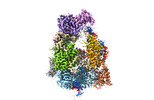 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
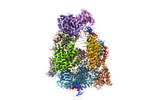
| Title | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationfollicle cell of egg chamber stalk formation / regulatory ncRNA processing / bidentate ribonuclease III activity / positive regulation of Toll signaling pathway / MicroRNA (miRNA) biogenesis / Small interfering RNA (siRNA) biogenesis / PKR-mediated signaling / dsRNA transport / regulation of regulatory ncRNA processing / dosage compensation by hyperactivation of X chromosome ...follicle cell of egg chamber stalk formation / regulatory ncRNA processing / bidentate ribonuclease III activity / positive regulation of Toll signaling pathway / MicroRNA (miRNA) biogenesis / Small interfering RNA (siRNA) biogenesis / PKR-mediated signaling / dsRNA transport / regulation of regulatory ncRNA processing / dosage compensation by hyperactivation of X chromosome / global gene silencing by mRNA cleavage /  ribonuclease III / ribonuclease III /  deoxyribonuclease I activity / deoxyribonuclease I activity /  apoptotic DNA fragmentation / detection of virus / RISC-loading complex / regulatory ncRNA-mediated post-transcriptional gene silencing / RISC complex assembly / apoptotic DNA fragmentation / detection of virus / RISC-loading complex / regulatory ncRNA-mediated post-transcriptional gene silencing / RISC complex assembly /  ribonuclease III activity / siRNA processing / siRNA binding / ATP-dependent activity, acting on RNA / positive regulation of innate immune response / RISC complex / heterochromatin formation / positive regulation of defense response to virus by host / locomotory behavior / mRNA 3'-UTR binding / ribonuclease III activity / siRNA processing / siRNA binding / ATP-dependent activity, acting on RNA / positive regulation of innate immune response / RISC complex / heterochromatin formation / positive regulation of defense response to virus by host / locomotory behavior / mRNA 3'-UTR binding /  helicase activity / cytoplasmic ribonucleoprotein granule / cellular response to virus / helicase activity / cytoplasmic ribonucleoprotein granule / cellular response to virus /  double-stranded RNA binding / defense response to virus / perinuclear region of cytoplasm / double-stranded RNA binding / defense response to virus / perinuclear region of cytoplasm /  ATP hydrolysis activity / ATP hydrolysis activity /  RNA binding / RNA binding /  ATP binding / ATP binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) | |||||||||
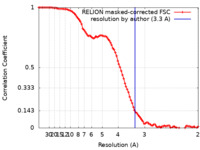
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Yamaguchi S / Nishizawa T / Kusakizako T / Yamashita K / Tomita A / Hirano H / Nishimasu H / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex. Authors: Sonomi Yamaguchi / Masahiro Naganuma / Tomohiro Nishizawa / Tsukasa Kusakizako / Yukihide Tomari / Hiroshi Nishimasu / Osamu Nureki /  Abstract: In flies, Argonaute2 (Ago2) and small interfering RNA (siRNA) form an RNA-induced silencing complex to repress viral transcripts. The RNase III enzyme Dicer-2 associates with its partner protein R2D2 ...In flies, Argonaute2 (Ago2) and small interfering RNA (siRNA) form an RNA-induced silencing complex to repress viral transcripts. The RNase III enzyme Dicer-2 associates with its partner protein R2D2 and cleaves long double-stranded RNAs to produce 21-nucleotide siRNA duplexes, which are then loaded into Ago2 in a defined orientation. Here we report cryo-electron microscopy structures of the Dicer-2-R2D2 and Dicer-2-R2D2-siRNA complexes. R2D2 interacts with the helicase domain and the central linker of Dicer-2 to inhibit the promiscuous processing of microRNA precursors by Dicer-2. Notably, our structure represents the strand-selection state in the siRNA-loading process, and reveals that R2D2 asymmetrically recognizes the end of the siRNA duplex with the higher base-pairing stability, and the other end is exposed to the solvent and is accessible by Ago2. Our findings explain how R2D2 senses the thermodynamic asymmetry of the siRNA and facilitates the siRNA loading into Ago2 in a defined orientation, thereby determining which strand of the siRNA duplex is used by Ago2 as the guide strand for target silencing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31742.map.gz emd_31742.map.gz | 5.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31742-v30.xml emd-31742-v30.xml emd-31742.xml emd-31742.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31742_fsc.xml emd_31742_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_31742.png emd_31742.png | 74.1 KB | ||
| Masks |  emd_31742_msk_1.map emd_31742_msk_1.map | 36.3 MB |  Mask map Mask map | |
| Others |  emd_31742_half_map_1.map.gz emd_31742_half_map_1.map.gz emd_31742_half_map_2.map.gz emd_31742_half_map_2.map.gz | 32.5 MB 32.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31742 http://ftp.pdbj.org/pub/emdb/structures/EMD-31742 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31742 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31742 | HTTPS FTP |
-Related structure data
| Related structure data |  7v6cMC  7v6bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11099 (Title: Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex EMPIAR-11099 (Title: Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplexData size: 1.4 TB Data #1: Structure of the Dicer-2-R2D2 heterodimer [micrographs - multiframe] Data #2: Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31742.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31742.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.9985 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31742_msk_1.map emd_31742_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_31742_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_31742_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dicer-2-R2D2-siRNA
| Entire | Name: Dicer-2-R2D2-siRNA |
|---|---|
| Components |
|
-Supramolecule #1: Dicer-2-R2D2-siRNA
| Supramolecule | Name: Dicer-2-R2D2-siRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Macromolecule #1: Dicer-2, isoform A
| Macromolecule | Name: Dicer-2, isoform A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  deoxyribonuclease I deoxyribonuclease I |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 199.309 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: HHHHHHHHAA AMEDVEIKPR GYQLRLVDHL TKSNGIVYLP TGSGKTFVAI LVLKRFSQDF DKPIESGGKR ALFMCNTVEL ARQQAMAVR RCTNFKVGFY VGEQGVDDWT RGMWSDEIKK NQVLVGTAQV FLDMVTQTYV ALSSLSVVII DECHHGTGHH P FREFMRLF ...String: HHHHHHHHAA AMEDVEIKPR GYQLRLVDHL TKSNGIVYLP TGSGKTFVAI LVLKRFSQDF DKPIESGGKR ALFMCNTVEL ARQQAMAVR RCTNFKVGFY VGEQGVDDWT RGMWSDEIKK NQVLVGTAQV FLDMVTQTYV ALSSLSVVII DECHHGTGHH P FREFMRLF TIANQTKLPR VVGLTGVLIK GNEITNVATK LKELEITYRG NIITVSDTKE LENVMLYATK PTEVMVSFPH QE QVLTVTR LISAEIEKFY VSLDLMNIGV QPIRRSKSLQ CLRDPSKKSF VKQLFNDFLY QMKEYGIYAA SIAIISLIVE FDI KRRQAE TLSVKLMHRT ALTLCEKIRH LLVQKLQDMT YDDDDDNVNT EEVIMNFSTP KVQRFLMSLK VSFADKDPKD ICCL VFVER RYTCKCIYGL LLNYIQSTPE LRNVLTPQFM VGRNNISPDF ESVLERKWQK SAIQQFRDGN ANLMICSSVL EEGID VQAC NHVFILDPVK TFNMYVQSKG RARTTEAKFV LFTADKEREK TIQQIYQYRK AHNDIAEYLK DRVLEKTEPE LYEIKG HFQ DDIDPFTNEN GAVLLPNNAL AILHRYCQTI PTDAFGFVIP WFHVLQEDER DRIFGVSAKG KHVISINMPV NCMLRDT IY SDPMDNVKTA KISAAFKACK VLYSLGELNE RFVPKTLKER VASIADVHFE HWNKYGDSVT ATVNKADKSK DRTYKTEC P LEFYDALPRV GEICYAYEIF LEPQFESCEY TEHMYLNLQT PRNYAILLRN KLPRLAEMPL FSNQGKLHVR VANAPLEVI IQNSEQLELL HQFHGMVFRD ILKIWHPFFV LDRRSKENSY LVVPLILGAG EQKCFDWELM TNFRRLPQSH GSNVQQREQQ PAPRPEDFE GKIVTQWYAN YDKPMLVTKV HRELTPLSYM EKNQQDKTYY EFTMSKYGNR IGDVVHKDKF MIEVRDLTEQ L TFYVHNRG KFNAKSKAKM KVILIPELCF NFNFPGDLWL KLIFLPSILN RMYFLLHAEA LRKRFNTYLN LHLLPFNGTD YM PRPLEID YSLKRNVDPL GNVIPTEDIE EPKSLLEPMP TKSIEASVAN LEITEFENPW QKYMEPVDLS RNLLSTYPVE LDY YYHFSV GNVCEMNEMD FEDKEYWAKN QFHMPTGNIY GNRTPAKTNA NVPALMPSKP TVRGKVKPLL ILQKTVSKEH ITPA EQGEF LAAITASSAA DVFDMERLEI LGDSFLKLSA TLYLASKYSD WNEGTLTEVK SKLVSNRNLL FCLIDADIPK TLNTI QFTP RYTWLPPGIS LPHNVLALWR ENPEFAKIIG PHNLRDLALG DEESLVKGNC SDINYNRFVE GCRANGQSFY AGADFS SEV NFCVGLVTIP NKVIADTLEA LLGVIVKNYG LQHAFKMLEY FKICRADIDK PLTQLLNLEL GGKKMRANVN TTEIDGF LI NHYYLEKNLG YTFKDRRYLL QALTHPSYPT NRITGSYQEL EFIGDAILDF LISAYIFENN TKMNPGALTD LRSALVNN T TLACICVRHR LHFFILAENA KLSEIISKFV NFQESQGHRV TNYVRILLEE ADVQPTPLDL DDELDMTELP HANKCISQE AEKGVPPKGE FNMSTNVDVP KALGDVLEAL IAAVYLDCRD LQRTWEVIFN LFEPELQEFT RKVPINHIRQ LVEHKHAKPV FSSPIVEGE TVMVSCQFTC MEKTIKVYGF GSNKDQAKLS AAKHALQQLS KCDA |
-Macromolecule #2: R2D2
| Macromolecule | Name: R2D2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 39.920363 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKHRYTSLY KKAGSAAAPF TMDNKSAVSA LQEFCARTQI NLPTYSFIPG EDGGYVCKVE LLEIEALGN GRSKRDAKHL AASNILRKIQ LLPGIHGLMK DSTVGDLDEE LTNLNRDMVK ELRDYCVRRE MPLPCIEVVQ Q SGTPSAPE ...String: MDYKDHDGDY KDHDIDYKDD DDKHRYTSLY KKAGSAAAPF TMDNKSAVSA LQEFCARTQI NLPTYSFIPG EDGGYVCKVE LLEIEALGN GRSKRDAKHL AASNILRKIQ LLPGIHGLMK DSTVGDLDEE LTNLNRDMVK ELRDYCVRRE MPLPCIEVVQ Q SGTPSAPE FVACCSVASI VRYGKSDKKK DARQRAAIEM LALISSNSDN LRPDQMQVAS TSKLKVVDME ESMEELEALR RK KFTTYWE LKEAGSVDHT GMRLCDRHNY FKNFYPTLKK EAIEAINSDE YESSKDKAMD VMSSLKITPK ISEVESSSLV PLL SVELNC AFDVVLMAKE TDIYDHIIDY FRTMLI |
-Macromolecule #3: RNA (5'-R(P*UP*GP*AP*GP*G)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*GP*AP*GP*G)-3') / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 6.812045 KDa |
| Sequence | String: UGAGGUAGUA GGUUGUAUAG U |
-Macromolecule #4: RNA (5'-R(P*CP*CP*UP*CP*UP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*CP*CP*UP*CP*UP*CP*U)-3') / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 6.514892 KDa |
| Sequence | String: UAUACAACCU ACUACCUCUC U |
-Macromolecule #5: RNA (5'-R(*AP*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*...
| Macromolecule | Name: RNA (5'-R(*AP*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*G)-3') type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 7.141251 KDa |
| Sequence | String: AUGAGGUAGU AGGUUGUAUA GU |
-Macromolecule #6: RNA (5'-R(*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*...
| Macromolecule | Name: RNA (5'-R(*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*UP*C)-3') type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Drosophila melanogaster (fruit fly) Drosophila melanogaster (fruit fly) |
| Molecular weight | Theoretical: 6.820074 KDa |
| Sequence | String: CUAUACAACC UACUACCUCU CU |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X