[English] 日本語
 Yorodumi
Yorodumi- EMDB-28059: Venezuelan equine encephalitis virus-like particle in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
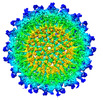
| Title | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT20 | |||||||||
 Map data Map data | Final map after post-processing | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Venezuelan equine encephalitis virus / Venezuelan equine encephalitis virus /  neutralizing antibody / VEEV / VIRUS LIKE PARTICLE-IMMUNE SYSTEM complex neutralizing antibody / VEEV / VIRUS LIKE PARTICLE-IMMUNE SYSTEM complex | |||||||||
| Biological species |   Venezuelan equine encephalitis virus / Venezuelan equine encephalitis virus /   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.7 Å cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Tsybovsky Y / Pletnev S / Verardi R / Roederer M / Kwong PD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Vaccine elicitation and structural basis for antibody protection against alphaviruses. Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / ...Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / Raffaello Verardi / Baoshan Zhang / David Ambrozak / Margaret Beddall / Hong Lei / Eun Sung Yang / Tracy Liu / Amy R Henry / Reda Rawi / Arne Schön / Chaim A Schramm / Chen-Hsiang Shen / Wei Shi / Tyler Stephens / Yongping Yang / Maria Burgos Florez / Julie E Ledgerwood / Crystal W Burke / Lawrence Shapiro / Julie M Fox / Peter D Kwong / Mario Roederer /  Abstract: Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine ...Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine encephalitis virus-like particles (VLPs), a regimen that protects against aerosol challenge with all three viruses. Single- and triple-virus-specific antibodies were isolated, and we identified 21 unique binding groups. Cryo-EM structures revealed that broad VLP binding inversely correlated with sequence and conformational variability. One triple-specific antibody, SKT05, bound proximal to the fusion peptide and neutralized all three Env-pseudotyped encephalitic alphaviruses by using different symmetry elements for recognition across VLPs. Neutralization in other assays (e.g., chimeric Sindbis virus) yielded variable results. SKT05 bound backbone atoms of sequence-diverse residues, enabling broad recognition despite sequence variability; accordingly, SKT05 protected mice against Venezuelan equine encephalitis virus, chikungunya virus, and Ross River virus challenges. Thus, a single vaccine-elicited antibody can protect in vivo against a broad range of alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28059.map.gz emd_28059.map.gz | 374.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28059-v30.xml emd-28059-v30.xml emd-28059.xml emd-28059.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28059_fsc.xml emd_28059_fsc.xml | 28.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_28059.png emd_28059.png | 101.5 KB | ||
| Masks |  emd_28059_msk_1.map emd_28059_msk_1.map | 1.9 GB |  Mask map Mask map | |
| Others |  emd_28059_additional_1.map.gz emd_28059_additional_1.map.gz emd_28059_half_map_1.map.gz emd_28059_half_map_1.map.gz emd_28059_half_map_2.map.gz emd_28059_half_map_2.map.gz | 1.5 GB 1.6 GB 1.6 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28059 http://ftp.pdbj.org/pub/emdb/structures/EMD-28059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28059 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28059 | HTTPS FTP |
-Related structure data
| Related structure data |  8decC  8dedC  8deeC  8defC  8deqC  8derC  8dulC  8dunC  8dwoC  8eeuC  8eevC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28059.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28059.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map after post-processing | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2315 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28059_msk_1.map emd_28059_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map before post-processing
| File | emd_28059_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map before post-processing | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2
| File | emd_28059_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1
| File | emd_28059_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between Venezuelan equine encephalitis virus-like particl...
| Entire | Name: Complex between Venezuelan equine encephalitis virus-like particle and Fab SKT20 |
|---|---|
| Components |
|
-Supramolecule #1: Complex between Venezuelan equine encephalitis virus-like particl...
| Supramolecule | Name: Complex between Venezuelan equine encephalitis virus-like particle and Fab SKT20 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Molecular weight | Theoretical: 33 MDa |
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Venezuelan equine encephalitis virus Venezuelan equine encephalitis virus |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKE ASQKQKGGGQ GKKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTF PIMLEGKING YACVVGGKLF RPMHVEGKID NDVLAALKTK ...String: MFPFQPMYPM QPMPYRNPFA APRRPWFPRT DPFLAMQVQE LTRSMANLTF KQRRDAPPEG PSAKKPKKE ASQKQKGGGQ GKKKKNQGKK KAKTGPPNPK AQNGNKKKTN KKPGKRQRMV MKLESDKTF PIMLEGKING YACVVGGKLF RPMHVEGKID NDVLAALKTK KASKYDLEYA DV PQNMRAD TFKYTHEKPQ GYYSWHHGAV QYENGRFTVP KGVGAKGDSG RPILDNQGRV VAI VLGGVN EGSRTALSVV MWNEKGVTVK YTPENCEQW SLVTTMCLLA NVTFPCAQPP ICYDR KPAE TLAMLSVNVD NPGYDELLEA AVKCPGRKRR STEELFNEY KLTRPYMARC IRCAVGS CH SPIAIEAVKS DGHDGYVRLQ TSSQYGLDSS GNLKGRTMRY DMHGTIKEIP LHQVSLYT S RPCHIVDGHG YFLLARCPAG DSITMEFKKD SVRHSCSVPY EVKFNPVGRE LYTHPPEHG VEQACQVYAH DAQNRGAYVE MHLPGSEVDS SLVSLSGSSV TVTPPDGTSA LVECECGGTK ISETINKTK QFSQCTKKEQ CRAYRLQNDK WVYNSDKLPK AAGATLKGKL HVPFLLADGK C TVPLAPEP MITFGFRSVS LKLHPKNPTY LITRQLADEP HYTHELISEP AVRNFTVTEK GW EFVWGNH PPKRFWAQET APGNPHGLPH EVITHYYHRY PMSTILGLSI CAAIATVSVA AST WLFCRS RVACLTPYRL TPNARIPFCL AVLCCARTAR AETTWESLDH LWNNNQQMFW IQLL IPLAA LIVVTRLLRC VCCVVPFLVM AGAAAPA YE HATTMPSQAG ISYNTIVNRA GYAPLP ISI TPTKIKLIPT VNLEYVTCHY KTGMDSPAIK CCGSQECTPT YRPDEQCKVF TGVYPFM WG GAYCFCDTEN TQVSKAYVMK SDDCLADHAE AYKAHTASVQ AFLNITVGEH SIVTTVYV N GETPVNFNGV KITAGPLSTA WTPFDRKIVQ YAGEIYNYDF PEYGAGQPGA FGDIQSRTV SSSDLYANTN LVLQRPKAGA IHVPYTQAPS GFEQWKKDKA PSLKFTAPFG CEIYTNPIRA ENCAVGSIP LAFDIPDALF TRVSETPTLS AAECTLNECV YSSDFGGIAT VKYSASKSGK C AVHVPSGT ATLKEAAVEL TEQGSATIHF STANIHPEFR LQICTSYVTC KGDCHPPKDH IV THPQYHA QTFTAAVSKT AWTWLTSLLG GSAVIIIIGL VLATIVAMYV LTNQKHN |
-Macromolecule #2: Fab SKT20 heavy chain
| Macromolecule | Name: Fab SKT20 heavy chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQVQESGPG LVKPSETLSL TCAVSSGSIN DDSYYWTWIR QSPGKGLEWL GFIHGGTGKS FYNPSLESRV TISKDTSRNQ FSLTLSSVSA ADTAVYYCAR SHFCSNTFCY GWFDVWGPGI RVTVSS AST KGPSVFPLAP SSRSTSESTA ALGCLVKDYF PEPVTVSWNS ...String: QVQVQESGPG LVKPSETLSL TCAVSSGSIN DDSYYWTWIR QSPGKGLEWL GFIHGGTGKS FYNPSLESRV TISKDTSRNQ FSLTLSSVSA ADTAVYYCAR SHFCSNTFCY GWFDVWGPGI RVTVSS AST KGPSVFPLAP SSRSTSESTA ALGCLVKDYF PEPVTVSWNS GSLTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYVC NVNHKPSNTK VDKRVEIKTC Gglevlfq |
-Macromolecule #3: Fab SKT20 light chain
| Macromolecule | Name: Fab SKT20 light chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVVMTQTPLS LPITPGEPAS ISCRSSQSLL HSNGNTYLHW YLQKPGQSPQ LLIYGGSNRA SGVPDRFSGS GSGTDFTLKI SKVEAEDVGV YYCVQAIAFP WTFGQGTKVE IK RTVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGAL KTGNSQESVT ...String: DVVMTQTPLS LPITPGEPAS ISCRSSQSLL HSNGNTYLHW YLQKPGQSPQ LLIYGGSNRA SGVPDRFSGS GSGTDFTLKI SKVEAEDVGV YYCVQAIAFP WTFGQGTKVE IK RTVAAPS VFIFPPSEDQ VKSGTVSVVC LLNNFYPREA SVKWKVDGAL KTGNSQESVT EQDSKDNTYS LSSTLTLSST EYQSHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm Bright-field microscopy / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 47.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z
Z Y
Y X
X


































