[English] 日本語
 Yorodumi
Yorodumi- EMDB-27723: Cryo-EM Structure of Antibody SKW11 in complex with Western Equin... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
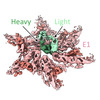
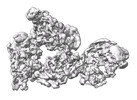
| Title | Cryo-EM Structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus spike (local refinement from VLP particles) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Antibody / SKW11 / Antibody / SKW11 /  Western Equine Encephalitis Virus / broadly neutralizing / Western Equine Encephalitis Virus / broadly neutralizing /  IMMUNE SYSTEM IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology information togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell ... togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell ... togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / togavirin / T=4 icosahedral viral capsid / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / symbiont entry into host cell / symbiont-mediated suppression of host gene expression / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / host cell nucleus / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity /  proteolysis / proteolysis /  RNA binding / RNA binding /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Western equine encephalitis virus / Western equine encephalitis virus /   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 5.84 Å cryo EM / Resolution: 5.84 Å | |||||||||
 Authors Authors | Cerutti G / Verardi R / Roederer M / Shapiro L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Vaccine elicitation and structural basis for antibody protection against alphaviruses. Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / ...Authors: Matthew S Sutton / Sergei Pletnev / Victoria Callahan / Sungyoul Ko / Yaroslav Tsybovsky / Tatsiana Bylund / Ryan G Casner / Gabriele Cerutti / Christina L Gardner / Veronica Guirguis / Raffaello Verardi / Baoshan Zhang / David Ambrozak / Margaret Beddall / Hong Lei / Eun Sung Yang / Tracy Liu / Amy R Henry / Reda Rawi / Arne Schön / Chaim A Schramm / Chen-Hsiang Shen / Wei Shi / Tyler Stephens / Yongping Yang / Maria Burgos Florez / Julie E Ledgerwood / Crystal W Burke / Lawrence Shapiro / Julie M Fox / Peter D Kwong / Mario Roederer /  Abstract: Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine ...Alphaviruses are RNA viruses that represent emerging public health threats. To identify protective antibodies, we immunized macaques with a mixture of western, eastern, and Venezuelan equine encephalitis virus-like particles (VLPs), a regimen that protects against aerosol challenge with all three viruses. Single- and triple-virus-specific antibodies were isolated, and we identified 21 unique binding groups. Cryo-EM structures revealed that broad VLP binding inversely correlated with sequence and conformational variability. One triple-specific antibody, SKT05, bound proximal to the fusion peptide and neutralized all three Env-pseudotyped encephalitic alphaviruses by using different symmetry elements for recognition across VLPs. Neutralization in other assays (e.g., chimeric Sindbis virus) yielded variable results. SKT05 bound backbone atoms of sequence-diverse residues, enabling broad recognition despite sequence variability; accordingly, SKT05 protected mice against Venezuelan equine encephalitis virus, chikungunya virus, and Ross River virus challenges. Thus, a single vaccine-elicited antibody can protect in vivo against a broad range of alphaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27723.map.gz emd_27723.map.gz | 1.1 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27723-v30.xml emd-27723-v30.xml emd-27723.xml emd-27723.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27723.png emd_27723.png | 143.2 KB | ||
| Others |  emd_27723_half_map_1.map.gz emd_27723_half_map_1.map.gz emd_27723_half_map_2.map.gz emd_27723_half_map_2.map.gz | 1 GB 1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27723 http://ftp.pdbj.org/pub/emdb/structures/EMD-27723 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27723 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27723 | HTTPS FTP |
-Related structure data
| Related structure data |  8dunMC  8decC  8dedC  8deeC  8defC  8deqC  8derC  8dulC  8dwoC  8eeuC  8eevC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27723.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27723.map.gz / Format: CCP4 / Size: 1.1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.66 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27723_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27723_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Western Equine Encephalitis Virus trimer in complex with Antibody...
| Entire | Name: Western Equine Encephalitis Virus trimer in complex with Antibody Fab SKW11 |
|---|---|
| Components |
|
-Supramolecule #1: Western Equine Encephalitis Virus trimer in complex with Antibody...
| Supramolecule | Name: Western Equine Encephalitis Virus trimer in complex with Antibody Fab SKW11 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Western equine encephalitis virus Western equine encephalitis virus |
-Macromolecule #1: Antibody SKW11 heavy chain
| Macromolecule | Name: Antibody SKW11 heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) |
| Molecular weight | Theoretical: 24.709627 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCAVSGDSIS NNYWTWIRHF PGKGLEWIGR IYGDAGSTDY NPSLKSRVTI SMDLSKNQFS LDLTSVTVA DTALYYCASR TTVADNWFDV WGPGVLVTVS SASTKGPSVF PLAPSSRSTS ESTAALGCLV KDYFPEPVTV S WNSGSLTS ...String: QVQLQESGPG LVKPSETLSL TCAVSGDSIS NNYWTWIRHF PGKGLEWIGR IYGDAGSTDY NPSLKSRVTI SMDLSKNQFS LDLTSVTVA DTALYYCASR TTVADNWFDV WGPGVLVTVS SASTKGPSVF PLAPSSRSTS ESTAALGCLV KDYFPEPVTV S WNSGSLTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYVCNVNHK PSNTKVDKRV EIKTCGGLEV LFQ |
-Macromolecule #2: Antibody SKW11 light chain
| Macromolecule | Name: Antibody SKW11 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Macaca fascicularis (crab-eating macaque) Macaca fascicularis (crab-eating macaque) |
| Molecular weight | Theoretical: 23.317848 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDTVT ITCRATQSIS SLLAWYQYKP GKAPKLLIYQ ASSLHIGVPS RFSGSGSGTD FTLTISSLQS EDFATYYCQ QHDSRPWTFG QGTKVDIKRT VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGALKTGNS Q ESVTEQDS ...String: DIQMTQSPSS LSASVGDTVT ITCRATQSIS SLLAWYQYKP GKAPKLLIYQ ASSLHIGVPS RFSGSGSGTD FTLTISSLQS EDFATYYCQ QHDSRPWTFG QGTKVDIKRT VAAPSVFIFP PSEDQVKSGT VSVVCLLNNF YPREASVKWK VDGALKTGNS Q ESVTEQDS KDNTYSLSST LTLSSTEYQS HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Western equine encephalitis virus Western equine encephalitis virus |
| Molecular weight | Theoretical: 52.905348 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RPTNAETFGE TLNHLWFNNQ PFLWAQLCIP LAALVILFRC FSCCMPFLLV AGVCLGKVDA FEHATTVPNV PGIPYKALVE RAGYAPLNL EITVVSSELT PSTNKEYVTC RFHTVIPSPQ VKCCGSLECK ASSKADYTCR VFGGVYPFMW GGAQCFCDSE N TQLSEAYV ...String: RPTNAETFGE TLNHLWFNNQ PFLWAQLCIP LAALVILFRC FSCCMPFLLV AGVCLGKVDA FEHATTVPNV PGIPYKALVE RAGYAPLNL EITVVSSELT PSTNKEYVTC RFHTVIPSPQ VKCCGSLECK ASSKADYTCR VFGGVYPFMW GGAQCFCDSE N TQLSEAYV EFAPDCTIDH AVALKVHTAA LKVGLRIVYG NTTAHLDTFV NGVTPGSSRD LKVIAGPISA AFSPFDHKVV IR KGLVYNY DFPEYGAMKP GAFGDIQASS LDATDIVART DIRLLKPSVK NIHVPYTQAV SGYEMWKNNS GRPLQETAPF GCK IEVEPL RASNCAYGHI PISIDIPDAA FVRSSESPTI LEVSCTVADC IYSADFGGSL TLQYKADREG HCPVHSHSTT AVLK EATTH VTAVGSITLH FSTSSPQANF IVSLCGKKTT CNAECKPPAD HIIGEPHKVD QEFQAAVSKT SWNWLLALFG GASSL IVVG LIVLV |
-Macromolecule #4: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Western equine encephalitis virus Western equine encephalitis virus |
| Molecular weight | Theoretical: 45.29373 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PYLGFCPYCR HSAPCFSPIK IENVWDESDD GSIRIQVSAQ FGYNQAGTAD VTKFRYMSFD HDHDIKEDSM DKIAISTSGP CRRLGHKGY FLLAQCPPGD SVTVSITSGA SENSCTVEKK IRRKFVGREE YLFPPVHGKL VKCHVYDHLK ETSAGYITMH R PGPHAYKS ...String: PYLGFCPYCR HSAPCFSPIK IENVWDESDD GSIRIQVSAQ FGYNQAGTAD VTKFRYMSFD HDHDIKEDSM DKIAISTSGP CRRLGHKGY FLLAQCPPGD SVTVSITSGA SENSCTVEKK IRRKFVGREE YLFPPVHGKL VKCHVYDHLK ETSAGYITMH R PGPHAYKS YLEEASGEVY IKPPSGKNVT YECKCGDYST GIVSTRTKMN GCTKAKQCIA YKSDQTKWVF NSPDLIRHTD HS VQGKLHI PFRLTPTVCP VPLAHTPTVT KWFKGITLHL TATRPTLLTT RKLGLRADAT AEWITGTTSR NFSVGREGLE YVW GNHEPV RVWAQESAPG DPHGWPHEII IHYYHRHPVY TVIVLCGVAL AILVGTASSA ACIAKARRDC LTPYALAPNA TVPT ALAVL CCI |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 46.5 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 5.84 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 854520 |
 Movie
Movie Controller
Controller





















 Z
Z Y
Y X
X

















