+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24470 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of precleavage Cre tetrameric complex | |||||||||
 Map data Map data | Precleavage synaptic complex of Cre recombinase mutant in complex with duplex loxP DNA | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Escherichia phage P1 (virus) Escherichia phage P1 (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.23 Å cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Stachowski K / Foster MP | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Mechanisms of Cre recombinase synaptic complex assembly and activation illuminated by Cryo-EM. Authors: Kye Stachowski / Andrew S Norris / Devante Potter / Vicki H Wysocki / Mark P Foster /  Abstract: Cre recombinase selectively recognizes DNA and prevents non-specific DNA cleavage through an orchestrated series of assembly intermediates. Cre recombines two loxP DNA sequences featuring a pair of ...Cre recombinase selectively recognizes DNA and prevents non-specific DNA cleavage through an orchestrated series of assembly intermediates. Cre recombines two loxP DNA sequences featuring a pair of palindromic recombinase binding elements and an asymmetric spacer region, by assembly of a tetrameric synaptic complex, cleavage of an opposing pair of strands, and formation of a Holliday junction intermediate. We used Cre and loxP variants to isolate the monomeric Cre-loxP (54 kDa), dimeric Cre2-loxP (110 kDa), and tetrameric Cre4-loxP2 assembly intermediates, and determined their structures using cryo-EM to resolutions of 3.9, 4.5 and 3.2 Å, respectively. Progressive and asymmetric bending of the spacer region along the assembly pathway enables formation of increasingly intimate interfaces between Cre protomers and illuminates the structural bases of biased loxP strand cleavage order and half-the-sites activity. Application of 3D variability analysis to the tetramer data reveals constrained conformational sampling along the pathway between protomer activation and Holliday junction isomerization. These findings underscore the importance of protein and DNA flexibility in Cre-mediated site selection, controlled activation of alternating protomers, the basis for biased strand cleavage order, and recombination efficiency. Such considerations may advance development of site-specific recombinases for use in gene editing applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24470.map.gz emd_24470.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24470-v30.xml emd-24470-v30.xml emd-24470.xml emd-24470.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24470.png emd_24470.png | 75 KB | ||
| Masks |  emd_24470_msk_1.map emd_24470_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_24470_half_map_1.map.gz emd_24470_half_map_1.map.gz emd_24470_half_map_2.map.gz emd_24470_half_map_2.map.gz | 59.4 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24470 http://ftp.pdbj.org/pub/emdb/structures/EMD-24470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24470 | HTTPS FTP |
-Related structure data
| Related structure data |  7rhxMC  7rhyC  7rhzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24470.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24470.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Precleavage synaptic complex of Cre recombinase mutant in complex with duplex loxP DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.899 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24470_msk_1.map emd_24470_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
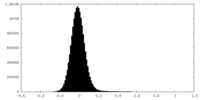
| Density Histograms |
-Half map: #2
| File | emd_24470_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
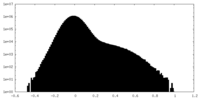
| Density Histograms |
-Half map: #1
| File | emd_24470_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Precleavage synaptic complex of Cre recombinase mutant K201A and ...
| Entire | Name: Precleavage synaptic complex of Cre recombinase mutant K201A and loxP DNA |
|---|---|
| Components |
|
-Supramolecule #1: Precleavage synaptic complex of Cre recombinase mutant K201A and ...
| Supramolecule | Name: Precleavage synaptic complex of Cre recombinase mutant K201A and loxP DNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Molecular weight | Theoretical: 54 KDa |
-Macromolecule #1: Recombinase cre
| Macromolecule | Name: Recombinase cre / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Molecular weight | Theoretical: 38.537062 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MSNLLTVHQN LPALPVDATS DEVRKNLMDM FRDRQAFSEH TWKMLLSVCR SWAAWCKLNN RKWFPAEPED VRDYLLYLQA RGLAVKTIQ QHLGQLNMLH RRSGLPRPSD SNAVSLVMRR IRKENVDAGE RAKQALAFER TDFDQVRSLM ENSDRCQDIR N LAFLGIAY ...String: MSNLLTVHQN LPALPVDATS DEVRKNLMDM FRDRQAFSEH TWKMLLSVCR SWAAWCKLNN RKWFPAEPED VRDYLLYLQA RGLAVKTIQ QHLGQLNMLH RRSGLPRPSD SNAVSLVMRR IRKENVDAGE RAKQALAFER TDFDQVRSLM ENSDRCQDIR N LAFLGIAY NTLLRIAEIA RIRVKDISRT DGGRMLIHIG RTATLVSTAG VEKALSLGVT KLVERWISVS GVADDPNNYL FC RVRKNGV AAPSATSQLS TRALEGIFEA THRLIYGAKD DSGQRYLAWS GHSARVGAAR DMARAGVSIP EIMQAGGWTN VNI VMNYIR NLDSETGAMV RLLEDGD |
-Macromolecule #2: DNA (42-MER)
| Macromolecule | Name: DNA (42-MER) / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Molecular weight | Theoretical: 12.833278 KDa |
| Sequence | String: (DC)(DC)(DG)(DC)(DA)(DT)(DA)(DA)(DC)(DT) (DT)(DC)(DG)(DT)(DA)(DT)(DA)(DG)(DC)(DA) (DT)(DA)(DC)(DA)(DT)(DT)(DA)(DT)(DA) (DC)(DG)(DA)(DA)(DG)(DT)(DT)(DA)(DT)(DC) (DG) (DC)(DC) |
-Macromolecule #3: DNA (42-MER)
| Macromolecule | Name: DNA (42-MER) / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia phage P1 (virus) Escherichia phage P1 (virus) |
| Molecular weight | Theoretical: 13.024384 KDa |
| Sequence | String: (DG)(DG)(DC)(DG)(DA)(DT)(DA)(DA)(DC)(DT) (DT)(DC)(DG)(DT)(DA)(DT)(DA)(DA)(DT)(DG) (DT)(DA)(DT)(DG)(DC)(DT)(DA)(DT)(DA) (DC)(DG)(DA)(DA)(DG)(DT)(DT)(DA)(DT)(DG) (DC) (DG)(DG) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: Buffer was made fresh from solid reagents and filtered with a 0.22 um filter. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector with two hexapoles. Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 86.0 K / Max: 86.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3662 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 / Details: 45 total frames |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 737980 |
|---|---|
| CTF correction | Software - Name: cryoSPARC (ver. 3.0.1) |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.0.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.0.1) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.23 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.0.1) / Number images used: 315574 |
 Movie
Movie Controller
Controller

















 Z
Z Y
Y X
X