+ Open data
Open data
- Basic information
Basic information
| Entry | 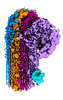 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
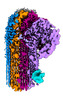
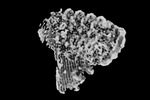
| Title | CryoEM reconstruction of C5b8-CD59, density subtracted | |||||||||
 Map data Map data | CryoEM reconstruction of C5b8-CD59. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Bubeck D / Couves EC / Gardner S | |||||||||
| Funding support | European Union,  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for membrane attack complex inhibition by CD59. Authors: Emma C Couves / Scott Gardner / Tomas B Voisin / Jasmine K Bickel / Phillip J Stansfeld / Edward W Tate / Doryen Bubeck /  Abstract: CD59 is an abundant immuno-regulatory receptor that protects human cells from damage during complement activation. Here we show how the receptor binds complement proteins C8 and C9 at the membrane to ...CD59 is an abundant immuno-regulatory receptor that protects human cells from damage during complement activation. Here we show how the receptor binds complement proteins C8 and C9 at the membrane to prevent insertion and polymerization of membrane attack complex (MAC) pores. We present cryo-electron microscopy structures of two inhibited MAC precursors known as C5b8 and C5b9. We discover that in both complexes, CD59 binds the pore-forming β-hairpins of C8 to form an intermolecular β-sheet that prevents membrane perforation. While bound to C8, CD59 deflects the cascading C9 β-hairpins, rerouting their trajectory into the membrane. Preventing insertion of C9 restricts structural transitions of subsequent monomers and indirectly halts MAC polymerization. We combine our structural data with cellular assays and molecular dynamics simulations to explain how the membrane environment impacts the dual roles of CD59 in controlling pore formation of MAC, and as a target of bacterial virulence factors which hijack CD59 to lyse human cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15800.map.gz emd_15800.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15800-v30.xml emd-15800-v30.xml emd-15800.xml emd-15800.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15800_fsc.xml emd_15800_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15800.png emd_15800.png | 117.4 KB | ||
| Masks |  emd_15800_msk_1.map emd_15800_msk_1.map | 343 MB |  Mask map Mask map | |
| Others |  emd_15800_half_map_1.map.gz emd_15800_half_map_1.map.gz emd_15800_half_map_2.map.gz emd_15800_half_map_2.map.gz | 318.6 MB 318.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15800 http://ftp.pdbj.org/pub/emdb/structures/EMD-15800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15800 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15800.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15800.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of C5b8-CD59. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.171 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15800_msk_1.map emd_15800_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
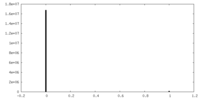
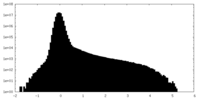
| Density Histograms |
-Half map: CryoEM reconstruction of C5b8-CD59.
| File | emd_15800_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of C5b8-CD59. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM reconstruction of C5b8-CD59.
| File | emd_15800_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of C5b8-CD59. | ||||||||||||
| Projections & Slices |
| ||||||||||||
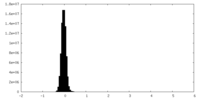
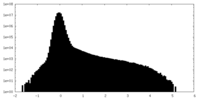
| Density Histograms |
- Sample components
Sample components
-Entire : C5b8-CD59
| Entire | Name: C5b8-CD59 |
|---|---|
| Components |
|
-Supramolecule #1: C5b8-CD59
| Supramolecule | Name: C5b8-CD59 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#7 / Details: Inhibited complement membrane attack complex |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number real images: 12805 / Average exposure time: 9.8 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Initial rigid body fits were performed using UCSF Chimera and UCSF ChimeraX. Carbohydrates were added using Coot. Models were refined iteratively using Isolde and Phenix Real Space Refine. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 87 |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X