+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | NuA4 Histone Acetyltransferase complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NuA4 /  histone acetyltransferase complex / histone acetyltransferase complex /  Epigenetics / Epigenetics /  DNA repair / DNA repair /  TRANSCRIPTION TRANSCRIPTION | |||||||||
| Biological species |  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Frechard A / Hexnerova R / Crucifix C / Papai G / Smirnova E / Faux C / Lo Ying Ping F / Helmlinger D / Schultz P / Ben-shem A | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification. Authors: Alexander Fréchard / Céline Faux / Rozalie Hexnerova / Corinne Crucifix / Gabor Papai / Ekaterina Smirnova / Conor McKeon / Florie Lo Ying Ping / Dominique Helmlinger / Patrick Schultz / Adam Ben-Shem /  Abstract: Histone acetylation regulates most DNA transactions and is dynamically controlled by highly conserved enzymes. The only essential histone acetyltransferase (HAT) in yeast, Esa1, is part of the 1-MDa ...Histone acetylation regulates most DNA transactions and is dynamically controlled by highly conserved enzymes. The only essential histone acetyltransferase (HAT) in yeast, Esa1, is part of the 1-MDa NuA4 complex, which plays pivotal roles in both transcription and DNA-damage repair. NuA4 has the unique capacity to acetylate histone targets located several nucleosomes away from its recruitment site. Neither the molecular mechanism of this activity nor its physiological importance are known. Here we report the structure of the Pichia pastoris NuA4 complex, with its core resolved at 3.4-Å resolution. Three subunits, Epl1, Eaf1 and Swc4, intertwine to form a stable platform that coordinates all other modules. The HAT module is firmly anchored into the core while retaining the ability to stretch out over a long distance. We provide structural, biochemical and genetic evidence that an unfolded linker region of the Epl1 subunit is critical for this long-range activity. Specifically, shortening the Epl1 linker causes severe growth defects and reduced H4 acetylation levels over broad chromatin regions in fission yeast. Our work lays the foundations for a mechanistic understanding of NuA4's regulatory role and elucidates how its essential long-range activity is attained. #1:  Journal: Res Sq / Year: 2022 Journal: Res Sq / Year: 2022Title: The structure of the NuA4/Tip60 complex reveals the mechanism and importance of long-range chromatin modification Authors: Schultz P / Frechard A / Hexnerova R / Crucifix C / Papai G / Smirnova E / Faux C / Ping F / Helmlinger D / Ben-Shem A | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14980.map.gz emd_14980.map.gz | 15.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14980-v30.xml emd-14980-v30.xml emd-14980.xml emd-14980.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14980_fsc.xml emd_14980_fsc.xml | 17.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14980.png emd_14980.png | 57.9 KB | ||
| Masks |  emd_14980_msk_1.map emd_14980_msk_1.map | 343 MB |  Mask map Mask map | |
| Others |  emd_14980_half_map_1.map.gz emd_14980_half_map_1.map.gz emd_14980_half_map_2.map.gz emd_14980_half_map_2.map.gz | 318.6 MB 318.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14980 http://ftp.pdbj.org/pub/emdb/structures/EMD-14980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14980 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14980.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14980.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.862 Å | ||||||||||||||||||||||||||||||||||||
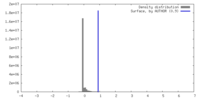
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14980_msk_1.map emd_14980_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
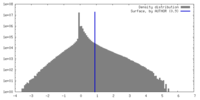
| Density Histograms |
-Half map: #1
| File | emd_14980_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
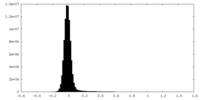
| Density Histograms |
-Half map: #2
| File | emd_14980_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
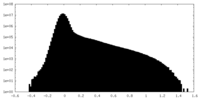
| Density Histograms |
- Sample components
Sample components
-Entire : NuA4 histone acetyltransferase complex
| Entire | Name: NuA4 histone acetyltransferase complex |
|---|---|
| Components |
|
-Supramolecule #1: NuA4 histone acetyltransferase complex
| Supramolecule | Name: NuA4 histone acetyltransferase complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
-Macromolecule #1: Tra1
| Macromolecule | Name: Tra1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MLHVVQLDDF ATRLKAAEDY QSKHSVLSEI CDSLETFNAA QDYEYFLKSL IPLFIDVLKE VPVSFVANS PENKLRNITL EILHRIPAND ALQAYSNEIV DTLMDLLKVE NELNGILCMK A ITTLHKTF KASLQEKVHP FIDIVIEIYS NIPQVVEEQF NGNQIDSKEN ...String: MLHVVQLDDF ATRLKAAEDY QSKHSVLSEI CDSLETFNAA QDYEYFLKSL IPLFIDVLKE VPVSFVANS PENKLRNITL EILHRIPAND ALQAYSNEIV DTLMDLLKVE NELNGILCMK A ITTLHKTF KASLQEKVHP FIDIVIEIYS NIPQVVEEQF NGNQIDSKEN VDSTSRPNSP SF SSQSDDS KQLAQAMFSF KTLAESPITM VSLYSSYKEL AASSLGNFIP HVMKVLSLEV AKQ AEARKA AEEKGIILVN VCKEITNRAN YGEFIIGQVK AASFLAYLFI RRQAQTFLEP YQQA IPDII IRLLQDCPSE LSAARKELLH ATRHILSTDF RKMFIPKIDL LFDLRVLIGE GFTAY ETLR PLAYSTVADF IHNVRDHLTP AQLWKSVSIY CKNLQDDSLA LTVQIMSAKL LLNLIE KIM RSESKTESRQ LLMVIIDAYT KRFKMLNSRY NGIMKQHATY EKEKQEKQNQ ERLLTNK LD GTTPSPSDDK KVELIDEDQD VKMEDPTPEI SDQETIKGDN DASTEPQDSE QQLADFMS L QEYLPIQVSV PPEIDLLKDS RYLFKTLMTF LKTIMIGLKN SNPPSSQNHF NAQNWNETA RGFSNEDINI LKSLFRECIL ALRFFSTSKT SLPASSMKQS FDITGPNLPI TSTKEEKDLM EIFATMFIH IDPASFNEIV REELPFMYKQ MLDFASLLHI PQFFLASVIT SSSFSGILIT F LKSKLVDL GEVNIIKSNI LIRLFKLCFM SVSLFPAANE SVILPHLNEL ILKSLKLSTT AK EPLVYFY LIRTLFRSIG GGRFENLYKE IMPLLQVLLE SLSKLIHEAR RPQERDIYVE LCL TVPVRL SVLVPHLSYL MKPLVYALNG SQESVSQGLR TLELCVDNLT AEYFDPIIEP VIDD VMEAL SKHLKPLPYY HQHSHTTLRI LGKLGGRNRT FIKPVDNLKT DSELFQNVEA MFKIH GLPN EVPLSITPGL SAAFSLLTDP RPRIHYRINS FKYISGIFQL FLGATQLPDD YANRLK ESM DIILEDTIAP DEPLNKLHHF PVKDIAKYDS QMELLVKLLE SIFYAVSLQE VREESKA LI RGTCNHFILL YFNKMVIDKR KFVRKFSVDN HEGNLFLNEN CIFDAIIYAL SSDNSAVR S MGLESVQLIY DSCVELFGNI DCALKFAPLN VMCSKFIHCC FEEPYHKKLA GCIGLEMML NSLDIPMKYF NARQLEIIRA LFYVLRDTAP ELPCEVTNTA KRLILNSLKE WNKELTRNDV FSSVFQNLV SSLIVDLPNA NEIVRATAQE ALRTLSETTQ VPIATMISPC KHILLAPIFG K PLRALPFQ MQIGNIDAIT FCMGLENSFL EYNEELNRLV QEALALVDAE DESLVSAHRI SE HKTSEQL VRLRVVCIQL LSLAITKPEF AAAQQRSNIR VKILVVFFKS LCGRSIEIIR AAH GGLKAV IDLKMKLPKE LLQNGLRPML MNLSDHKKLT VASLEALSGL LKLFISYFKV GIGS KLLDH LLAWAQPRTL QQLGSQDLEN NSTVQIIVAI LDVFHLLPPT AHKFMNDLMN ALLYL ENNL HRCQYSPFRE PLAKFLDRFP DESFEYFFNE FSKREITTRF VYFVGLDSCS SLRAKV LES LPRVRGLLHQ EGSAEEKCVR FSNLVDLCES LAASDKEWIK DKEELLGELL DAGSVCL TL KRSSNVVSPL YFQVDQGFET LQLLYIEYFK SQPLGHEKVF NFIDKISKEG LPFVLEFD D FIFNEVVKCQ DIPTVQQTLD TIIRMTPQVS SLDARVYLYK RIFLPICIYE SEMHGDLSR LSQTENNELP AWLKSFDSDV WKATGPLVDD YTSTLEDRYR LELMQLTALL IKGAPTALTD MRKDIIKFS WNYIKLDDNT SKQAAYVVTA YFISRFDTPS ELTTRIFVAL LRCHQIDTRY L VKQALELL APVLSERTNS ELDWLKWPRR VLSEDGFNIT QVANIYQLIV KFPDLFYPAR DH FIPNIIT AMGKLTVMSN TSLENQQLAI DLAELILKWE TKLPKSEKLG SAEETEKEKS VSE DKMDID VKEETKEDIA ERPKAEDQIG GDDSDSSNIL TSEDYEVSFA QREACVTFLI RYIC ISTQR PSENELGKRA LNILYELLGP KYWSEVTVKL QFFERFLMSS DLNQPSLLGY CLNAL EVLA VALKWKPTTW IIENVSYLQK LLEKCLRSDN QDIQEILQKV LGIILEAINK ETQGSE EDE PEEVTNFISL IVNIIGEDLS NMTSVAAGVS LCWTLSLYRP NALDSLLPSI MRTFNKL CR DHIAISLQGN QPQSGDFANI EFEAKVTTNL LEKILNLCAA RISSLDDQRR VFLSLLAQ L IDRSVDKDML LKVINIVTEW IFKTDFYPTT KEKAGILGKM MIFDLRGEPE LSKKFNQVI VDIFESKELA HTELTARMET AFLFGTRLSD VSIRKKLMSI LSDSLELDID KRLFYIIKDQ NWEYLSDYP WLNQALQLLY GSFHLDSPIR LSPEENTLSP LQSITEGLAR EKSPVEKAPQ N IIDFVAKH NEFLDSVRSL TAGDILNPLI DISYQSAETI HNAWVVVFPV AYSAIESRYE LE FTRALVK LLFKDYHIRQ QDARPNVIKS LLDGVGKCPG LHLPPHLVKY LGSNYNAWYG AIK LLEELS EGQGIDNQKI SDANQDALLE VYMSLQEDDM FYGTWRRRAK YFETNAALSY EQIG IWDKA LQLYEAAQIK ARSGVFPFGE SEYSLWEDHW IYCAEKLQHW EILTELAKHE GFTDL LLEC GWRGADWIAD REPLEQSVKT VMDIPTPRRQ IFQTFLALQG FSQQKDTLQD VSRLCD EGI QLTLRKWNAL PQRVTRAHIG LLHTFQQYVE LMEASQVYSS LVTTNAQNLD VKSQELK RV LQAWRERLPN VWDDINIWND LVTWRQHVFG VINRVYMPFV PVLQQSNGTN NGNSYAYR G YHEMAWVINR FAHVARKHEM PEVCINQLTK IYTLPNIEIQ EAFLKLREQA KCHYQNSSE LNTGLDVISN TNLVYFATQQ KAEFFTLKGM FLAKLNAKDE ANQAFATAVQ IDLNLPKAWA EWGFFNDRR FKENPEEIFH AKNAISCYLQ AAGLYKDGKT RKLLCRILWL ISLDDAAGSL A KTFEDHHG ESPVWYWITF VPQLLTSLSH KEAKIVRHIL IQIAKSYPQS LHFQLRTTKE DY QAIQRQA MAVNRAEEQS SNKQDTADSV LKNTNTPQPQ TRTETSGTTA ESDKKPSIPP KEE QGSPQP SRPATTQASP QAQSQENGES SQKHPPEIPT TDSRQPWQDV EEIMGILKTA YPLL ALSLE SLVDQLNQRF KCNADEDAYR LVIVLYNDGV QQMNRVANPR EEVKLPAATE ASISR FADS VLPKNIREVF EQDIIACNPN LETYISKLRK WRDCLEEKLD RSYGKADLER VSLHLS LFH HQKFEDIEIP GQYLLHKDNN NHFIKIERFL PTLDLVRGSN GCYKRMTIRG NDGSLHP FA VQFPAARHCR REERIFQLFR IFDDALSRKV QSRRRNISLT LPIAVPLSPH IRILNDDK R YTTLMGIYEE FCRRKGQSRD EPFAYTIQKL RAAFDPRLPK PDIVSVRAEV LASIQSTLV PSTLLKDYYT EKFSNYENYW LFRKQFTAQY ASFIFMTYIM CINSRQPQKI HINEGSGNIW TSEMLPTKV ATGKTHSTAY NNSTLDPAVK AGAPIFYNTE SVPFRLTPNI QKFIGEAGLE G ILSVYILV IANSLSDSEF DMEQYLSLFV RDEVISWFAQ QHRASAQTNQ LREIVRVNVE LL TKRVLQL NHIPNSQNVA TQFVLNLISQ AVNPRNLAYT DSAWMAYL |
-Macromolecule #2: Epl1
| Macromolecule | Name: Epl1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MVLPSAAGAR FRQRKISVKQ TLQVFKQSDI ADLETEDQQR ELQKIETGVE KGEEEEEHLQ RVINAAQAA VISGGKVEKA YIPTPDASQV WKDYDKYYRD KFQPPGTYIR FSATVEETTG C IYNLDEED ESFLKEVLNK NLANGIKPCT ETELEIVLQR FEVVIDKKQP ...String: MVLPSAAGAR FRQRKISVKQ TLQVFKQSDI ADLETEDQQR ELQKIETGVE KGEEEEEHLQ RVINAAQAA VISGGKVEKA YIPTPDASQV WKDYDKYYRD KFQPPGTYIR FSATVEETTG C IYNLDEED ESFLKEVLNK NLANGIKPCT ETELEIVLQR FEVVIDKKQP FLAMDPSQIL TF EELHSAA LVVDPNSIEE IELSLEKQLG LHPFRTLLDG QRSQLSHRKL AELLRIFGQK IYD HWKQRR IARHGRPITA QLRFEDSSEK DDSDPYVCFR RREFRQARKT RRTDTQGSER LRKL HRELK QTRDLLLAVA QREVKRKEAI EVGHEVFQLR CSVKTLKRDL GVKGEEEDLV AHKRK KVVP TTDEDRKYRK GYNSGNYPNR NQAQAQQQQQ QLQQQQISKS GLNPVSIQPY VKLPMS RIP DMDLITVSTV LNEKDEAINR AVAEKLRQRK ESDRGWVNLT DDPFNPFLQF TNPDSIL EK GHFPYSSIAA ALFEVDQSNY FDPEITQLIK DKKPLPRTLC FKDNALTTPL PPSIYEVA S NNKLDVTAPI CKVRKRMGRR GLWIDRKMTV DEPLDEFLDF STFEKSLDAD ITDRNSAKS QQGMNVYDSV DDANSRLRSR FSFDRDVPLF NPVDPSELNQ ISSQTQSIRF GCMLLTKAYE QVHQAKQKL FLEQQQHQQK QQQKLLQQRQ KVQQKLQQQQ QQQQQAQQQN PKSNTNKVVN Q INNTKSPV KQSKIPSKVS KNISGVTTSA GKGA |
-Macromolecule #3: Swc4
| Macromolecule | Name: Swc4 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MSSDILDVLS ISGRSNLSNQ KKKITKDGPT KKKKQTAMSR ELFNLIGQNT PPLAVEKTVK FKEKLNVNN KPTPWSYVEF SNDARESKDG LKLHHWIKGS SELAKNSPYL FEKYNQKIQI P SFTKEEYD EFLKDLDLEC REREKRDKAL KEQEAKEAEK LEKEKENDKE ...String: MSSDILDVLS ISGRSNLSNQ KKKITKDGPT KKKKQTAMSR ELFNLIGQNT PPLAVEKTVK FKEKLNVNN KPTPWSYVEF SNDARESKDG LKLHHWIKGS SELAKNSPYL FEKYNQKIQI P SFTKEEYD EFLKDLDLEC REREKRDKAL KEQEAKEAEK LEKEKENDKE RVNGDELGKE ED NASDFVP KKENVNDQIL PKIDEPQTNK KDDMESTEDV SKSDSQEVSK DVNPSEEVEA SWD YDETVH LFQLCEKWDL RWPIIVDRYE YDERSMEELK ERFYKVSERI LRHKYRNVTM DDKT SLLVQ TLSSFDKRRE TERKQYLRRL LSRSPTEIAE EESLVIEARK FELAAKKMLT ERASL LRLL DSPQSTGSIS QYLTSQGLTQ LYNTLMSADR SKRRKVETPT PPQIPPGASS SLHRTS MDL KRKAAKKIGP NALLENIKAT GNSVPPSGNA PQSAAIELIN TKMTPEEKEA YGIKIHQ EK LQPGVSLRSA RLPTFKPATQ AKIVVVLNEL EVSPKPTIPT AKVVAQYDNL LQTINVLL E TKKQVNKLEV ELNLLEGKEE DKES |
-Macromolecule #4: Arp4
| Macromolecule | Name: Arp4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MATAPQVYGA DEINAVVLDA SSYRTKIGYG SLDCPILNLP SYYGHQTKDG SEKFIFEENS MLIPRPDYE IKKIMKDGII DDFEGAVKQY NYMFDVLKLK PSEQPILVIE STQNEYEKKT A LLKQLLKE NKFVATFLIK NPTCVSFAHG RPNCLVVDLG HDLVTITPIL ...String: MATAPQVYGA DEINAVVLDA SSYRTKIGYG SLDCPILNLP SYYGHQTKDG SEKFIFEENS MLIPRPDYE IKKIMKDGII DDFEGAVKQY NYMFDVLKLK PSEQPILVIE STQNEYEKKT A LLKQLLKE NKFVATFLIK NPTCVSFAHG RPNCLVVDLG HDLVTITPIL DGISLRKQVL GT HYAGAFL SQQLRQLLNH KGVEVVPVYK VKSKVPTYFP DEAKFEERKY DFDISESFEN FHK LRILRE MKETLLQALP DSETEKLKEQ ETEEDTRYFE FPNGLNVPFT KYERVRLANS LFNP SEPYT GESGPNIVVE GFSVETGKII NEDTITSREY VPLRRSKKTD SSSGRRSKDE PTDDK PRGL TSLVNQALNH LDVDLKPQLA NNIILTGATS LIPGVAERLN QELTAMNPGL KVRIHS SAN VIERTCSAWI GGSILSSLGT FHQLWVSENE YDEVGAKKLI MDRFR |
-Macromolecule #5: Eaf1
| Macromolecule | Name: Eaf1 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MSSLDSNVTA SSKHDSPKRQ PEDGSGTNDD PLQRILNERK RKLAELYCVS RLPLLPISPS QVSQIGENL MRFLERNDLE QGRQFNITTL TGDKSQKQPP VSKEVSTADQ LESPHWPVKE D EETTKDEP PRKKQKTAST SAEPQKATHK ESKMTMMHQE VTESEAPTLS ...String: MSSLDSNVTA SSKHDSPKRQ PEDGSGTNDD PLQRILNERK RKLAELYCVS RLPLLPISPS QVSQIGENL MRFLERNDLE QGRQFNITTL TGDKSQKQPP VSKEVSTADQ LESPHWPVKE D EETTKDEP PRKKQKTAST SAEPQKATHK ESKMTMMHQE VTESEAPTLS SLIRKYAQDE VP IRPDDPT DRDLNFELLD RNKTIIQALP EIYPHKIADS ASLTELYYLT QTFPLAKLLP RSH KSLTTD AYESALLEGK IAVLYSRIEE LKRQRKWSLR QPKRFIDPFT RESPTHWDHL LAEM KWLSV DIMEERKFKA ASCVQLAQAV SDYWTYGKIV CIQRKPLIFL TDEEIKERTV TKVMK TEVD NENEHDHDDN DIFKDAQEEP RAMDQEQNQP FEENATIDVA KLLERPNPKD EIIPPA LPT YSMGDYKRLN QNAEPFKLHI GLDDFKKEDL VLVEKLPLSF IFDDNLSDSK KKLSEYE KA PIAAISTLLA PPEDDEWYKI VIRRDPASEL SASLDYQKGL FGASSQRRYN VLKPPKPP P IKNLELRTPT IWLPQDDKLL IRYVAEYAFN WDIISAHLSA RPARAYVANI ERRTPWQCF ERYIQLNDKF QFTDMRGQYA QSAQAWLEAA HKTQSTTKRR ISPLGVGIES IQRGHRRLRW GSMLDAMRK CMRRRENINR SSQVERKHTS DDKRTNVPTP EELSRLKYDR DKAIQEAYMH Q NSGTFSSA RPHTQSSALS PSKGKNAPLP TSAQQTGTHP YTNGIPPKGT LPKTTTPAPG VP STQPGTP NTRQPQVSSR VATSQNTTPV TNRTGTPNGN RPGPNNSFTQ EQLQHFLQAQ RHR QMMQTS AGTPVNKSNV PNRPAGITNT NVTTPANAVS SSTTPSKSAA TASQQQPSPS RGIN LTPAQ VNALINQIQA QNPNMSKDQV TKFAVAYIQN LQNQRERSSN GHSTPQVRTV PTPTR ATAG PPVSASPVTS NASPTTPASL TKEKLDALEN SPNLTPQQRQ QITLFKAMQA RNKMNQ VAN RGQESASASS NLSASPNTDT KQTTDTNK |
-Macromolecule #6: Act1
| Macromolecule | Name: Act1 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Sequence | String: MDGEDVAALV IDNGSGMCKA GYAGDDAPHT VFPSVVGRPR HQGVMVGMGQ KDSFVGDEAQ SKRGILTLR YPIEHGIVTN WDDMEKIWHH TFYNELRLAP EEHPVLLTEA PMNPKSNREK M TQIMFETF NVPAFYVSIQ AVLSLYASGR TTGIVLDSGD GVTHVVPIYA ...String: MDGEDVAALV IDNGSGMCKA GYAGDDAPHT VFPSVVGRPR HQGVMVGMGQ KDSFVGDEAQ SKRGILTLR YPIEHGIVTN WDDMEKIWHH TFYNELRLAP EEHPVLLTEA PMNPKSNREK M TQIMFETF NVPAFYVSIQ AVLSLYASGR TTGIVLDSGD GVTHVVPIYA GFSLPHAILR ID LAGRDLT DYLMKILSER GYTFSTSAER EIVRDIKEKL CYVALDFDQE LQTSSQSSSI EKS YELPDG QVITIGNERF RAPEALFHPS VLGLEASGID QTTYNSIMKC DVDVRKELYS NIVM SGGTT MFPGIAERMQ KELTALAPSS MKVKISAPPE RKYSVWIGGS ILASLGTFQQ MWISK QEYD ESGPSIVHLK CF |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 1.4000000000000001 µm Bright-field microscopy / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 1.4000000000000001 µm |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 52.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)