[English] 日本語
 Yorodumi
Yorodumi- EMDB-12632: Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12632 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound by Blasticidin S. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationL13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Formation of a pool of free 40S subunits / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Translation initiation complex formation / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition ...L13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Formation of a pool of free 40S subunits / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Translation initiation complex formation / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / Major pathway of rRNA processing in the nucleolus and cytosol / GTP hydrolysis and joining of the 60S ribosomal subunit / translation termination factor activity / Translesion synthesis by REV1 / Recognition of DNA damage by PCNA-containing replication complex / Translesion Synthesis by POLH / Downregulation of ERBB4 signaling / Spry regulation of FGF signaling / Downregulation of ERBB2:ERBB3 signaling / NOD1/2 Signaling Pathway / APC/C:Cdc20 mediated degradation of Cyclin B / SCF-beta-TrCP mediated degradation of Emi1 / APC-Cdc20 mediated degradation of Nek2A / EGFR downregulation / TCF dependent signaling in response to WNT / NRIF signals cell death from the nucleus / p75NTR recruits signalling complexes / NF-kB is activated and signals survival / Activated NOTCH1 Transmits Signal to the Nucleus / Downregulation of TGF-beta receptor signaling / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / Downregulation of SMAD2/3:SMAD4 transcriptional activity / SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription / Senescence-Associated Secretory Phenotype (SASP) / Regulation of innate immune responses to cytosolic DNA / activated TAK1 mediates p38 MAPK activation / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / Regulation of FZD by ubiquitination / PINK1-PRKN Mediated Mitophagy / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Regulation of TNFR1 signaling / TNFR1-induced NF-kappa-B signaling pathway / Translesion synthesis by POLK / Translesion synthesis by POLI / Regulation of necroptotic cell death / MAP3K8 (TPL2)-dependent MAPK1/3 activation / HDR through Homologous Recombination (HRR) / Josephin domain DUBs / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / DNA Damage Recognition in GG-NER / Formation of Incision Complex in GG-NER / Gap-filling DNA repair synthesis and ligation in GG-NER / Dual Incision in GG-NER / Fanconi Anemia Pathway / Regulation of TP53 Activity through Phosphorylation / Regulation of TP53 Degradation / Regulation of TP53 Activity through Methylation / Negative regulation of MET activity / Cyclin D associated events in G1 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Downregulation of ERBB2 signaling / E3 ubiquitin ligases ubiquitinate target proteins / Regulation of PTEN localization / ER Quality Control Compartment (ERQC) / Regulation of expression of SLITs and ROBOs / Interferon alpha/beta signaling / Endosomal Sorting Complex Required For Transport (ESCRT) / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / IRAK2 mediated activation of TAK1 complex / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Alpha-protein kinase 1 signaling pathway / RAS processing / Pexophagy / Inactivation of CSF3 (G-CSF) signaling / Negative regulation of FLT3 /  Regulation of BACH1 activity / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / Regulation of NF-kappa B signaling / Termination of translesion DNA synthesis / Ovarian tumor domain proteases / Negative regulators of DDX58/IFIH1 signaling / Negative regulation of FGFR1 signaling / Negative regulation of FGFR2 signaling / Negative regulation of FGFR3 signaling / Negative regulation of FGFR4 signaling / Negative regulation of MAPK pathway / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Iron uptake and transport / Deactivation of the beta-catenin transactivating complex / Metalloprotease DUBs / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Activation of NF-kappaB in B cells / L13a-mediated translational silencing of Ceruloplasmin expression / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / Cdc20:Phospho-APC/C mediated degradation of Cyclin A Regulation of BACH1 activity / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / Regulation of NF-kappa B signaling / Termination of translesion DNA synthesis / Ovarian tumor domain proteases / Negative regulators of DDX58/IFIH1 signaling / Negative regulation of FGFR1 signaling / Negative regulation of FGFR2 signaling / Negative regulation of FGFR3 signaling / Negative regulation of FGFR4 signaling / Negative regulation of MAPK pathway / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Iron uptake and transport / Deactivation of the beta-catenin transactivating complex / Metalloprotease DUBs / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Activation of NF-kappaB in B cells / L13a-mediated translational silencing of Ceruloplasmin expression / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 / Cdc20:Phospho-APC/C mediated degradation of Cyclin ASimilarity search - Function | |||||||||
| Biological species |   Oryctolagus cuniculus (rabbit) / Oryctolagus cuniculus (rabbit) /   Rabbit (rabbit) / Rabbit (rabbit) /   Bovine (cattle) Bovine (cattle) | |||||||||
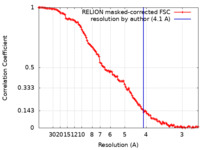
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.1 Å cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Powers KT / Yadav SKN / Bufton JC / Schaffitzel C | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA. Authors: Kyle T Powers / Flint Stevenson-Jones / Sathish K N Yadav / Beate Amthor / Joshua C Bufton / Ufuk Borucu / Dakang Shen / Jonas P Becker / Daria Lavysh / Matthias W Hentze / Andreas E Kulozik ...Authors: Kyle T Powers / Flint Stevenson-Jones / Sathish K N Yadav / Beate Amthor / Joshua C Bufton / Ufuk Borucu / Dakang Shen / Jonas P Becker / Daria Lavysh / Matthias W Hentze / Andreas E Kulozik / Gabriele Neu-Yilik / Christiane Schaffitzel /   Abstract: Deciphering translation is of paramount importance for the understanding of many diseases, and antibiotics played a pivotal role in this endeavour. Blasticidin S (BlaS) targets translation by binding ...Deciphering translation is of paramount importance for the understanding of many diseases, and antibiotics played a pivotal role in this endeavour. Blasticidin S (BlaS) targets translation by binding to the peptidyl transferase center of the large ribosomal subunit. Using biochemical, structural and cellular approaches, we show here that BlaS inhibits both translation elongation and termination in Mammalia. Bound to mammalian terminating ribosomes, BlaS distorts the 3'CCA tail of the P-site tRNA to a larger extent than previously reported for bacterial ribosomes, thus delaying both, peptide bond formation and peptidyl-tRNA hydrolysis. While BlaS does not inhibit stop codon recognition by the eukaryotic release factor 1 (eRF1), it interferes with eRF1's accommodation into the peptidyl transferase center and subsequent peptide release. In human cells, BlaS inhibits nonsense-mediated mRNA decay and, at subinhibitory concentrations, modulates translation dynamics at premature termination codons leading to enhanced protein production. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12632.map.gz emd_12632.map.gz | 15.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12632-v30.xml emd-12632-v30.xml emd-12632.xml emd-12632.xml | 98.5 KB 98.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12632_fsc.xml emd_12632_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12632.png emd_12632.png | 79.4 KB | ||
| Masks |  emd_12632_msk_1.map emd_12632_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_12632_half_map_1.map.gz emd_12632_half_map_1.map.gz emd_12632_half_map_2.map.gz emd_12632_half_map_2.map.gz | 80.9 MB 80.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12632 http://ftp.pdbj.org/pub/emdb/structures/EMD-12632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12632 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12632 | HTTPS FTP |
-Related structure data
| Related structure data |  7nwhMC  7nwgC  7nwiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12632.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12632.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12632_msk_1.map emd_12632_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12632_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12632_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound b...
+Supramolecule #1: Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound b...
+Macromolecule #1: 60S ribosomal protein L8
+Macromolecule #2: uL4
+Macromolecule #3: eL31
+Macromolecule #4: 40S ribosomal protein S3
+Macromolecule #5: S29
+Macromolecule #6: 60S ribosomal protein L5
+Macromolecule #7: Ribosomal protein L32
+Macromolecule #8: 40S ribosomal protein S4
+Macromolecule #9: 40S ribosomal protein S30
+Macromolecule #10: eL29
+Macromolecule #11: 60S ribosomal protein L6
+Macromolecule #12: eL33
+Macromolecule #13: Ribosomal protein S5
+Macromolecule #14: 40S ribosomal protein S27a
+Macromolecule #15: uL30
+Macromolecule #16: 60S ribosomal protein L34
+Macromolecule #17: 40S ribosomal protein S3a
+Macromolecule #18: 40S ribosomal protein S6
+Macromolecule #19: Epididymis tissue sperm binding protein Li 3a
+Macromolecule #20: L7a
+Macromolecule #21: uL29
+Macromolecule #22: S7
+Macromolecule #24: 40S ribosomal protein S27
+Macromolecule #25: L9
+Macromolecule #26: 60S ribosomal protein L36
+Macromolecule #27: 40S ribosomal protein S8
+Macromolecule #28: Eukaryotic peptide chain release factor subunit 1
+Macromolecule #29: 60S ribosomal protein L10
+Macromolecule #30: Ribosomal protein L37
+Macromolecule #31: 40S ribosomal protein S9
+Macromolecule #32: eRF3a
+Macromolecule #33: 60S ribosomal protein L11
+Macromolecule #34: L38
+Macromolecule #35: 40S ribosomal protein S10
+Macromolecule #36: 60S ribosomal protein L13
+Macromolecule #37: eL39
+Macromolecule #38: 40S ribosomal protein S11
+Macromolecule #39: Ribosomal protein L14
+Macromolecule #40: 60S RIBOSOMAL PROTEIN EL40
+Macromolecule #41: 40S ribosomal protein S12
+Macromolecule #42: Ribosomal protein L15
+Macromolecule #43: 60s ribosomal protein l41
+Macromolecule #44: ribosomal protein uS15
+Macromolecule #45: 60S RIBOSOMAL PROTEIN UL13
+Macromolecule #46: eL42
+Macromolecule #47: uS11
+Macromolecule #48: uL22
+Macromolecule #49: ribosomal protein eL43
+Macromolecule #50: 40S ribosomal protein uS19
+Macromolecule #51: eL18
+Macromolecule #52: eL28
+Macromolecule #53: Ribosomal protein S16
+Macromolecule #54: 60S ribosomal protein L19
+Macromolecule #55: 60S acidic ribosomal protein P0
+Macromolecule #56: 40S ribosomal protein S17
+Macromolecule #57: 60S ribosomal protein L18a
+Macromolecule #58: uL12
+Macromolecule #59: 40S ribosomal protein uS13
+Macromolecule #60: eL21
+Macromolecule #61: eS19
+Macromolecule #62: L22
+Macromolecule #63: 40S ribosomal protein S20
+Macromolecule #64: eL14
+Macromolecule #65: 40S ribosomal protein S21
+Macromolecule #66: 60S ribosomal protein L24-like protein
+Macromolecule #68: Ribosomal protein S15a
+Macromolecule #69: uL23
+Macromolecule #71: 40S ribosomal protein uS12
+Macromolecule #72: Ribosomal protein L26
+Macromolecule #74: 40S ribosomal protein S24
+Macromolecule #75: 60S ribosomal protein L27
+Macromolecule #77: 40S ribosomal protein S25
+Macromolecule #78: 60S ribosomal protein L27a
+Macromolecule #79: 40S_SA_C domain-containing protein
+Macromolecule #80: 40S ribosomal protein S26
+Macromolecule #81: uL3
+Macromolecule #82: eL30
+Macromolecule #83: 40S ribosomal protein S2
+Macromolecule #84: 40S ribosomal protein S28
+Macromolecule #23: mRNA
+Macromolecule #67: 28S Ribosomal RNA
+Macromolecule #70: 5S Ribosomal RNA
+Macromolecule #73: 5.8S Ribosomal RNA
+Macromolecule #76: 18S Ribosomal RNA
+Macromolecule #85: ZINC ION
+Macromolecule #86: MAGNESIUM ION
+Macromolecule #87: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
+Macromolecule #88: BLASTICIDIN S
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.63 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM HEPES pH 7.4 100 mM KOAc 5 mM Mg(Oac)2 1mM DTT |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 288.15 K / Instrument: LEICA EM GP Details: 30s sample incubation on grid followed by 1.1s blotting time.. |
| Details | 165nM (OD260nm~10.5) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.4 µm / Calibrated magnification: 79000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Details | Collected in super-resolution mode (0.675A pixel size) |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3500 / Average exposure time: 8.0 sec. / Average electron dose: 41.92 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X