+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11992 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the molecular sociology at the HeLa cell nuclear periphery | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  electron tomography / electron tomography /  cryo EM cryo EM | |||||||||
 Authors Authors | Mahamid J / Pfeffer S / Schaffer M / Villa E / Danev R / Kuhn-Cuellar L / Foerster F / Hyman A / Plitzko J / Baumeister W | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Visualizing the molecular sociology at the HeLa cell nuclear periphery. Authors: Julia Mahamid / Stefan Pfeffer / Miroslava Schaffer / Elizabeth Villa / Radostin Danev / Luis Kuhn Cuellar / Friedrich Förster / Anthony A Hyman / Jürgen M Plitzko / Wolfgang Baumeister /   Abstract: The molecular organization of eukaryotic nuclear volumes remains largely unexplored. Here we combined recent developments in cryo-electron tomography (cryo-ET) to produce three-dimensional snapshots ...The molecular organization of eukaryotic nuclear volumes remains largely unexplored. Here we combined recent developments in cryo-electron tomography (cryo-ET) to produce three-dimensional snapshots of the HeLa cell nuclear periphery. Subtomogram averaging and classification of ribosomes revealed the native structure and organization of the cytoplasmic translation machinery. Analysis of a large dynamic structure-the nuclear pore complex-revealed variations detectable at the level of individual complexes. Cryo-ET was used to visualize previously elusive structures, such as nucleosome chains and the filaments of the nuclear lamina, in situ. Elucidation of the lamina structure provides insight into its contribution to metazoan nuclear stiffness. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11992.map.gz emd_11992.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11992-v30.xml emd-11992-v30.xml emd-11992.xml emd-11992.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11992.png emd_11992.png | 327.1 KB | ||
| Masks |  emd_11992_msk_1.map emd_11992_msk_1.map emd_11992_msk_2.map emd_11992_msk_2.map emd_11992_msk_3.map emd_11992_msk_3.map emd_11992_msk_4.map emd_11992_msk_4.map emd_11992_msk_5.map emd_11992_msk_5.map emd_11992_msk_6.map emd_11992_msk_6.map emd_11992_msk_7.map emd_11992_msk_7.map | 1.6 GB 1.6 GB 1.6 GB 821.3 MB 1.6 GB 1.6 GB 1.6 GB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11992 http://ftp.pdbj.org/pub/emdb/structures/EMD-11992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11992 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11992.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11992.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 16.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11992_msk_1.map emd_11992_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
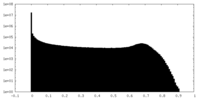
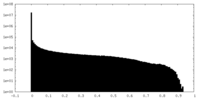
| Density Histograms |
-Mask #2
| File |  emd_11992_msk_2.map emd_11992_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
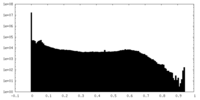
| Density Histograms |
-Mask #3
| File |  emd_11992_msk_3.map emd_11992_msk_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #4
| File |  emd_11992_msk_4.map emd_11992_msk_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #5
| File |  emd_11992_msk_5.map emd_11992_msk_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #6
| File |  emd_11992_msk_6.map emd_11992_msk_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #7
| File |  emd_11992_msk_7.map emd_11992_msk_7.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Interphase HeLa cell
| Entire | Name: Interphase HeLa cell |
|---|---|
| Components |
|
-Supramolecule #1: Interphase HeLa cell
| Supramolecule | Name: Interphase HeLa cell / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron tomography electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 0.5 nA / Focused ion beam - Duration: 1800 sec. / Focused ion beam - Temperature: 94 K / Focused ion beam - Initial thickness: 10000 nm / Focused ion beam - Final thickness: 190 nm Focused ion beam - Details: The value given for _emd_sectioning_focused_ion_beam.instrument is Quanta 3D FEG, FEI. This is not in a list of allowed values {'DB235', 'OTHER'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 0.8 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD / Number images used: 50 IMOD / Number images used: 50 |
|---|
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X