[English] 日本語
 Yorodumi
Yorodumi- SASDEK7: SpaO C-terminus (Surface presentation of antigens protein SpaO(SP... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  |
|---|---|
 Sample Sample | SpaO C-terminus
|
| Function / homology |  Type III secretion system apparatus protein YscQ/HrcQ/SpaO / Type III secretion system apparatus protein YscQ/HrcQ/SpaO /  Type III secretion system outer membrane, SpaO / Type III secretion system outer membrane, SpaO /  Flagellar motor switch protein FliN-like, C-terminal domain / SpoA-like superfamily / Type III flagellar switch regulator (C-ring) FliN C-term / bacterial-type flagellum-dependent swarming motility / Flagellar motor switch protein FliN-like, C-terminal domain / SpoA-like superfamily / Type III flagellar switch regulator (C-ring) FliN C-term / bacterial-type flagellum-dependent swarming motility /  protein secretion by the type III secretion system / positive chemotaxis / Surface presentation of antigens protein SpaO protein secretion by the type III secretion system / positive chemotaxis / Surface presentation of antigens protein SpaO Function and homology information Function and homology information |
| Biological species |   Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes. Authors: Ivonne Bernal / Jonathan Börnicke / Johannes Heidemann / Dmitri Svergun / Julia A Horstmann / Marc Erhardt / Anne Tuukkanen / Charlotte Uetrecht / Michael Kolbe /  Abstract: Many medically relevant Gram-negative bacteria use the type III secretion system (T3SS) to translocate effector proteins into the host for their invasion and intracellular survival. A multi-protein ...Many medically relevant Gram-negative bacteria use the type III secretion system (T3SS) to translocate effector proteins into the host for their invasion and intracellular survival. A multi-protein complex located at the cytosolic interface of the T3SS is proposed to act as a sorting platform by selecting and targeting substrates for secretion through the system. However, the precise stoichiometry and 3D organization of the sorting platform components are unknown. Here we reconstitute soluble complexes of the Salmonella Typhimurium sorting platform proteins including the ATPase InvC, the regulator OrgB, the protein SpaO and a recently identified subunit SpaO, which we show to be essential for the solubility of SpaO. We establish domain-domain interactions, determine for the first time the stoichiometry of each subunit within the complexes by native mass spectrometry and gain insight into their organization using small-angle X-ray scattering. Importantly, we find that in solution the assembly of SpaO/SpaO/OrgB/InvC adopts an extended L-shaped conformation resembling the sorting platform pods seen in in situ cryo-electron tomography, proposing that this complex is the core building block that can be conceivably assembled into higher oligomers to form the T3SS sorting platform. The determined molecular arrangements of the soluble complexes of the sorting platform provide important insights into its architecture and assembly. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-Models
- Sample
Sample
 Sample Sample | Name: SpaO C-terminus / Specimen concentration: 12.5 mg/ml |
|---|---|
| Buffer | Name: 20 mM HEPES pH 7.5, 150 mM NaCl / pH: 7.5 |
| Entity #835 | Name: SpaO C-terminus / Type: protein Description: Surface presentation of antigens protein SpaO(SPOA1,2) C-terminus Formula weight: 19 / Num. of mol.: 1 Source: Salmonella enterica subsp. enterica serovar Typhimurium References: UniProt: P40699 Sequence: MASWSHPQFE KGAVGGGRPK MLRWPLRFVI GSSDTQRSLL GRIGIGDVLL IRTSRAEVYC YAKKLGHFNR VEGGIIVETL DIQHIEEENN TTETAETLPG LNQLPVKLEF VLYRKNVTLA ELEAMGQQQL LSLPTNAELN VEIMANGVLL GNGELVQMND TLGVEIHEWL S |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm Synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||
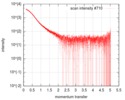
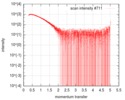
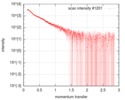
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result | Comments: SEC-SAXS was performed at 20°C using the following parameters: Column type, XXXX; Flow rate: XXXX mL/min; Total acquisition time: 60 min (3600 x 1 s frames); Injection concentration: XXXX ...Comments: SEC-SAXS was performed at 20°C using the following parameters: Column type, XXXX; Flow rate: XXXX mL/min; Total acquisition time: 60 min (3600 x 1 s frames); Injection concentration: XXXX mg/mL; Injection volume: XXXX μL.
|
 Movie
Movie Controller
Controller


 SASDEK7
SASDEK7