[English] 日本語
 Yorodumi
Yorodumi- SASDBG6: Ribosome biogenesis protein 15 (Nop15) (Ribosome biogenesis prote... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDBG6 |
|---|---|
 Sample Sample | Ribosome biogenesis protein 15 (Nop15)
|
| Function / homology |  Function and homology information Function and homology informationpreribosome, large subunit precursor / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) /  ribosomal large subunit biogenesis / ribosomal large subunit biogenesis /  rRNA binding / rRNA binding /  nucleolus / nucleolus /  RNA binding / RNA binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function |
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2017 Journal: Nucleic Acids Res / Year: 2017Title: Structural analysis reveals the flexible C-terminus of Nop15 undergoes rearrangement to recognize a pre-ribosomal RNA folding intermediate. Authors: Jun Zhang / Lauren E Gonzalez / Traci M Tanaka Hall /  Abstract: The RNA recognition motif (RRM) is the most abundant RNA-binding domain in eukaryotes, and it plays versatile roles in RNA metabolism. Despite its abundance, diversity of RRM structure and function ...The RNA recognition motif (RRM) is the most abundant RNA-binding domain in eukaryotes, and it plays versatile roles in RNA metabolism. Despite its abundance, diversity of RRM structure and function is generated by variations on a conserved core. Yeast Nop15 is an RRM protein that is essential for large ribosomal subunit biogenesis. We determined a 2.0 Å crystal structure of Nop15 that reveals a C-terminal α-helical region obscures its canonical RNA-binding surface. Small-angle X-ray scattering, NMR and RNA-binding analyses further reveal that the C-terminal residues of Nop15 are highly flexible, but essential for tight RNA binding. Moreover, comparison with a recently reported cryo-electron microscopy structure indicates that dramatic rearrangement of the C-terminal region of Nop15 in the pre-ribosome exposes the RNA-binding surface to recognize the base of its stem-loop target RNA and extends a newly-formed α helix to the distal loop where it forms protein interactions. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
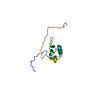
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #636 |  Type: mix / Software: EOM / Radius of dummy atoms: 1.90 A / Chi-square value: 0.649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #637 |  Type: mix / Software: EOM / Radius of dummy atoms: 1.90 A / Chi-square value: 0.649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #638 |  Type: mix / Software: EOM / Radius of dummy atoms: 1.90 A / Chi-square value: 0.649  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Ribosome biogenesis protein 15 (Nop15) / Specimen concentration: 0.70-3.00 / Specimen concentration: 0.70-3.00 |
|---|---|
| Buffer | Name: 25 mM HEPES, 500 mM NaCl, 2 mM DTT / pH: 7.5 |
| Entity #402 | Name: Nop15 / Type: protein / Description: Ribosome biogenesis protein 15 / Formula weight: 16.541 / Num. of mol.: 1 / Source: Saccharomyces cerevisiae / References: UniProt: P53927 / Formula weight: 16.541 / Num. of mol.: 1 / Source: Saccharomyces cerevisiae / References: UniProt: P53927Sequence: KDKKTLEEYS GIIYVSRLPH GFHEKELSKY FAQFGDLKEV RLARNKKTGN SRHYGFLEFV NKEDAMIAQE SMNNYLLMGH LLQVRVLPKG AKIEKLYKYK KRVLVEKGIT KPVKQLKDNM KQKHEERIKK LAKSGIEFKW |
-Experimental information
| Beam | Instrument name: Advanced Light Source (ALS) 12.3.1 (SIBYLS) City: Berkeley, CA / 国: USA  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.1 Å Synchrotron / Wavelength: 0.1 Å | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: MAR 165 CCD | |||||||||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller

 SASDBG6
SASDBG6