+Search query
-Structure paper
| Title | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 5899, Year 2023 |
| Publish date | Sep 22, 2023 |
 Authors Authors | Shota Suzuki / Kotaro Tanaka / Kouki Nishikawa / Hiroshi Suzuki / Atsunori Oshima / Yoshinori Fujiyoshi /  |
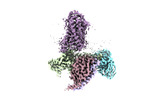
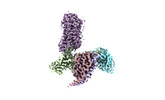
| PubMed Abstract | Hydroxycarboxylic acid receptors (HCA) are expressed in various tissues and immune cells. HCA2 and its agonist are thus important targets for treating inflammatory and metabolic disorders. Only ...Hydroxycarboxylic acid receptors (HCA) are expressed in various tissues and immune cells. HCA2 and its agonist are thus important targets for treating inflammatory and metabolic disorders. Only limited information is available, however, on the active-state binding of HCAs with agonists. Here, we present cryo-EM structures of human HCA2-Gi and HCA3-Gi signaling complexes binding with multiple compounds bound. Agonists were revealed to form a salt bridge with arginine, which is conserved in the HCA family, to activate these receptors. Extracellular regions of the receptors form a lid-like structure that covers the ligand-binding pocket. Although transmembrane (TM) 6 in HCAs undergoes dynamic conformational changes, ligands do not directly interact with amino acids in TM6, suggesting that indirect signaling induces a slight shift in TM6 to activate Gi proteins. Structural analyses of agonist-bound HCA2 and HCA3 together with mutagenesis and molecular dynamics simulation provide molecular insights into HCA ligand recognition and activation mechanisms. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37736747 / PubMed:37736747 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.85 - 3.21 Å |
| Structure data | EMDB-35442, PDB-8ihb: EMDB-35443, PDB-8ihf: EMDB-35444, PDB-8ihh: EMDB-35445, PDB-8ihi: EMDB-35446, PDB-8ihj: EMDB-35447, PDB-8ihk: |
| Chemicals |  ChemComp-NAG:  ChemComp-OKL:  ChemComp-FI7: 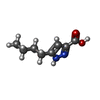 ChemComp-P8A: 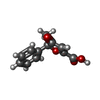 ChemComp-P9X: |
| Source |
|
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  GPCR GPCR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers