+Search query
-Structure paper
| Title | Molecular glue CELMoD compounds are regulators of cereblon conformation. |
|---|---|
| Journal, issue, pages | Science, Vol. 378, Issue 6619, Page 549-553, Year 2022 |
| Publish date | Nov 4, 2022 |
 Authors Authors | Edmond R Watson / Scott Novick / Mary E Matyskiela / Philip P Chamberlain / Andres H de la Peña / Jinyi Zhu / Eileen Tran / Patrick R Griffin / Ingrid E Wertz / Gabriel C Lander /  |
| PubMed Abstract | Cereblon (CRBN) is a ubiquitin ligase (E3) substrate receptor protein co-opted by CRBN E3 ligase modulatory drug (CELMoD) agents that target therapeutically relevant proteins for degradation. Prior ...Cereblon (CRBN) is a ubiquitin ligase (E3) substrate receptor protein co-opted by CRBN E3 ligase modulatory drug (CELMoD) agents that target therapeutically relevant proteins for degradation. Prior crystallographic studies defined the drug-binding site within CRBN's thalidomide-binding domain (TBD), but the allostery of drug-induced neosubstrate binding remains unclear. We performed cryo-electron microscopy analyses of the DNA damage-binding protein 1 (DDB1)-CRBN apo complex and compared these structures with DDB1-CRBN in the presence of CELMoD compounds alone and complexed with neosubstrates. Association of CELMoD compounds to the TBD is necessary and sufficient for triggering CRBN allosteric rearrangement from an open conformation to the canonical closed conformation. The neosubstrate Ikaros only stably associates with the closed CRBN conformation, illustrating the importance of allostery for CELMoD compound efficacy and informing structure-guided design strategies to improve therapeutic efficacy. |
 External links External links |  Science / Science /  PubMed:36378961 / PubMed:36378961 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.1 - 3.9 Å |
| Structure data | EMDB-27012, PDB-8cvp: EMDB-27234, PDB-8d7u: EMDB-27235, PDB-8d7v: EMDB-27236, PDB-8d7w: EMDB-27237, PDB-8d7x: EMDB-27238, PDB-8d7y:  EMDB-27239: Cereblon bound to CC-92480, Focused Refinement EMDB-27240, PDB-8d7z: EMDB-27241, PDB-8d80: EMDB-27242, PDB-8d81: |
| Chemicals |  ChemComp-ZN: 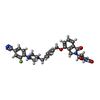 ChemComp-QFC: 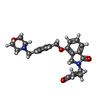 ChemComp-8W7: 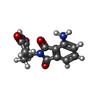 ChemComp-Y70: |
| Source |
|
 Keywords Keywords |  LIGASE / LIGASE /  Ubiquitin / Ubiquitin /  CRL4 / CRL4 /  Adaptor / Adaptor /  Cereblon / E3 Cereblon / E3 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers






















