+Search query
-Structure paper
| Title | Activation of Thoeris antiviral system via SIR2 effector filament assembly. |
|---|---|
| Journal, issue, pages | Nature, Vol. 627, Issue 8003, Page 431-436, Year 2024 |
| Publish date | Feb 21, 2024 |
 Authors Authors | Giedre Tamulaitiene / Dziugas Sabonis / Giedrius Sasnauskas / Audrone Ruksenaite / Arunas Silanskas / Carmel Avraham / Gal Ofir / Rotem Sorek / Mindaugas Zaremba / Virginijus Siksnys /   |
| PubMed Abstract | To survive bacteriophage (phage) infections, bacteria developed numerous anti-phage defence systems. Some of them (for example, type III CRISPR-Cas, CBASS, Pycsar and Thoeris) consist of two modules: ...To survive bacteriophage (phage) infections, bacteria developed numerous anti-phage defence systems. Some of them (for example, type III CRISPR-Cas, CBASS, Pycsar and Thoeris) consist of two modules: a sensor responsible for infection recognition and an effector that stops viral replication by destroying key cellular components. In the Thoeris system, a Toll/interleukin-1 receptor (TIR)-domain protein, ThsB, acts as a sensor that synthesizes an isomer of cyclic ADP ribose, 1''-3' glycocyclic ADP ribose (gcADPR), which is bound in the Smf/DprA-LOG (SLOG) domain of the ThsA effector and activates the silent information regulator 2 (SIR2)-domain-mediated hydrolysis of a key cell metabolite, NAD (refs. ). Although the structure of ThsA has been solved, the ThsA activation mechanism remained incompletely understood. Here we show that 1''-3' gcADPR, synthesized in vitro by the dimeric ThsB' protein, binds to the ThsA SLOG domain, thereby activating ThsA by triggering helical filament assembly of ThsA tetramers. The cryogenic electron microscopy (cryo-EM) structure of activated ThsA revealed that filament assembly stabilizes the active conformation of the ThsA SIR2 domain, enabling rapid NAD depletion. Furthermore, we demonstrate that filament formation enables a switch-like response of ThsA to the 1''-3' gcADPR signal. |
 External links External links |  Nature / Nature /  PubMed:38383786 PubMed:38383786 |
| Methods | EM (helical sym.) / X-ray diffraction |
| Resolution | 2.75 - 3.1 Å |
| Structure data | EMDB-16233, PDB-8bto: EMDB-16234, PDB-8btp:  PDB-8btn: |
| Chemicals | 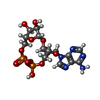 ChemComp-OJC:  ChemComp-NAD: 
ChemComp-RK3:  ChemComp-ENA: |
| Source |
|
 Keywords Keywords |  HYDROLASE / Thoeris / ThsB / HYDROLASE / Thoeris / ThsB /  TIR domain / TIR domain /  SIR2 domain / SIR2 domain /  SLOG domain / 3'cADPR SLOG domain / 3'cADPR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








