+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical structure of BcThsA in complex with 1''-3'gcADPR | |||||||||
 Map data Map data | Sharpened EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Thoeris /  SIR2 domain / SIR2 domain /  SLOG domain / 3'cADPR / SLOG domain / 3'cADPR /  HYDROLASE HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology information NAD+ glycohydrolase / defense response to virus / NAD+ glycohydrolase / defense response to virus /  hydrolase activity / hydrolase activity /  nucleotide binding / nucleotide binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Bacillus cereus (bacteria) / Bacillus cereus (bacteria) /   Bacillus cereus MSX-D12 (bacteria) Bacillus cereus MSX-D12 (bacteria) | |||||||||
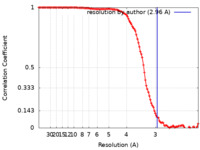
| Method | helical reconstruction /  cryo EM / Resolution: 2.96 Å cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Tamulaitiene G / Sasnauskas G / Sabonis D | |||||||||
| Funding support | Lithuania, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Activation of Thoeris antiviral system via SIR2 effector filament assembly. Authors: Giedre Tamulaitiene / Dziugas Sabonis / Giedrius Sasnauskas / Audrone Ruksenaite / Arunas Silanskas / Carmel Avraham / Gal Ofir / Rotem Sorek / Mindaugas Zaremba / Virginijus Siksnys /   Abstract: To survive bacteriophage (phage) infections, bacteria developed numerous anti-phage defence systems. Some of them (for example, type III CRISPR-Cas, CBASS, Pycsar and Thoeris) consist of two modules: ...To survive bacteriophage (phage) infections, bacteria developed numerous anti-phage defence systems. Some of them (for example, type III CRISPR-Cas, CBASS, Pycsar and Thoeris) consist of two modules: a sensor responsible for infection recognition and an effector that stops viral replication by destroying key cellular components. In the Thoeris system, a Toll/interleukin-1 receptor (TIR)-domain protein, ThsB, acts as a sensor that synthesizes an isomer of cyclic ADP ribose, 1''-3' glycocyclic ADP ribose (gcADPR), which is bound in the Smf/DprA-LOG (SLOG) domain of the ThsA effector and activates the silent information regulator 2 (SIR2)-domain-mediated hydrolysis of a key cell metabolite, NAD (refs. ). Although the structure of ThsA has been solved, the ThsA activation mechanism remained incompletely understood. Here we show that 1''-3' gcADPR, synthesized in vitro by the dimeric ThsB' protein, binds to the ThsA SLOG domain, thereby activating ThsA by triggering helical filament assembly of ThsA tetramers. The cryogenic electron microscopy (cryo-EM) structure of activated ThsA revealed that filament assembly stabilizes the active conformation of the ThsA SIR2 domain, enabling rapid NAD depletion. Furthermore, we demonstrate that filament formation enables a switch-like response of ThsA to the 1''-3' gcADPR signal. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16233.map.gz emd_16233.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16233-v30.xml emd-16233-v30.xml emd-16233.xml emd-16233.xml | 27.8 KB 27.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16233_fsc.xml emd_16233_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16233.png emd_16233.png | 85.3 KB | ||
| Masks |  emd_16233_msk_1.map emd_16233_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16233.cif.gz emd-16233.cif.gz | 6.6 KB | ||
| Others |  emd_16233_additional_1.map.gz emd_16233_additional_1.map.gz emd_16233_additional_2.map.gz emd_16233_additional_2.map.gz emd_16233_additional_3.map.gz emd_16233_additional_3.map.gz emd_16233_additional_4.map.gz emd_16233_additional_4.map.gz emd_16233_additional_5.map.gz emd_16233_additional_5.map.gz emd_16233_half_map_1.map.gz emd_16233_half_map_1.map.gz emd_16233_half_map_2.map.gz emd_16233_half_map_2.map.gz | 62.1 MB 117 MB 60.3 MB 114.9 MB 114.9 MB 115.9 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16233 http://ftp.pdbj.org/pub/emdb/structures/EMD-16233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16233 | HTTPS FTP |
-Related structure data
| Related structure data |  8btoMC  8btnC  8btpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16233.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16233.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16233_msk_1.map emd_16233_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened EM map
| File | emd_16233_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement (mask on A subunit) sharpened EM...
| File | emd_16233_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement (mask on A subunit) sharpened EM map resampled on main EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement (mask on A subunit) EM map resampled on main EM map
| File | emd_16233_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement (mask on A subunit) EM map resampled on main EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement (mask on A subunit) EM half...
| File | emd_16233_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement (mask on A subunit) EM half map resampled on main EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement (mask on A subunit) EM half...
| File | emd_16233_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement (mask on A subunit) EM half map resampled on main EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16233_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16233_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BcThsA in complex with 1''-3'gcADPR
| Entire | Name: BcThsA in complex with 1''-3'gcADPR |
|---|---|
| Components |
|
-Supramolecule #1: BcThsA in complex with 1''-3'gcADPR
| Supramolecule | Name: BcThsA in complex with 1''-3'gcADPR / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Bacillus cereus (bacteria) Bacillus cereus (bacteria) |
-Macromolecule #1: NAD(+) hydrolase ThsA
| Macromolecule | Name: NAD(+) hydrolase ThsA / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number:  NAD+ glycohydrolase NAD+ glycohydrolase |
|---|---|
| Source (natural) | Organism:   Bacillus cereus MSX-D12 (bacteria) Bacillus cereus MSX-D12 (bacteria) |
| Molecular weight | Theoretical: 55.417746 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SGGMKMNPIV ELFIKDFTKE VMEENAAIFA GAGLSMSVGY VSWAKLLEPI AQEIGLDVNK ENDLVSLAQY YCNENQGNRG RINQIILDE FSRKVDLTEN HKILARLPIH TYWTTAYDRL IEKALEEENK IADVKYTVKQ LATTKVKRDA VVYKMHGDVE H PSEAVLIK ...String: SGGMKMNPIV ELFIKDFTKE VMEENAAIFA GAGLSMSVGY VSWAKLLEPI AQEIGLDVNK ENDLVSLAQY YCNENQGNRG RINQIILDE FSRKVDLTEN HKILARLPIH TYWTTAYDRL IEKALEEENK IADVKYTVKQ LATTKVKRDA VVYKMHGDVE H PSEAVLIK DDYEKYSIKM DPYIKALSGD LVSKTFLFVG FSFTDPNLDY ILSRVRSAYE RDQRRHYCLI KKEERRPDEL EA DFEYRVR KQELFISDLS RFNIKTIVLN NYNEITEILQ RIENNIKTKT VFLSGSAVEY NHWETEHAEQ FIHQLSKELI RKD FNIVSG FGLGVGSFVI NGVLEELYMN QGTIDDDRLI LRPFPQGKKG EEQWDKYRRD MITRTGVSIF LYGNKIDKGQ VVKA KGVQS EFNISFEQNN YVVPVGATGY IAKDLWNKVN EEFETYYPGA DARMKKLFGE LNNEALSIEE LINTIIEFVE ILSN UniProtKB: NAD(+) hydrolase ThsA |
-Macromolecule #2: (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,...
| Macromolecule | Name: (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione type: ligand / ID: 2 / Number of copies: 12 / Formula: OJC |
|---|---|
| Molecular weight | Theoretical: 541.3 Da |
| Chemical component information | 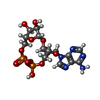 ChemComp-OJC: |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 12 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2321 / Average exposure time: 46.33 sec. / Average electron dose: 30.64 e/Å2 |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8bto: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)