+Search query
-Structure paper
| Title | Role of aIF5B in archaeal translation initiation. |
|---|---|
| Journal, issue, pages | Nucleic Acids Res, Vol. 50, Issue 11, Page 6532-6548, Year 2022 |
| Publish date | Jun 24, 2022 |
 Authors Authors | Ramy Kazan / Gabrielle Bourgeois / Christine Lazennec-Schurdevin / Eric Larquet / Yves Mechulam / Pierre-Damien Coureux / Emmanuelle Schmitt /  |
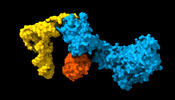
| PubMed Abstract | In eukaryotes and in archaea late steps of translation initiation involve the two initiation factors e/aIF5B and e/aIF1A. In eukaryotes, the role of eIF5B in ribosomal subunit joining is established ...In eukaryotes and in archaea late steps of translation initiation involve the two initiation factors e/aIF5B and e/aIF1A. In eukaryotes, the role of eIF5B in ribosomal subunit joining is established and structural data showing eIF5B bound to the full ribosome were obtained. To achieve its function, eIF5B collaborates with eIF1A. However, structural data illustrating how these two factors interact on the small ribosomal subunit have long been awaited. The role of the archaeal counterparts, aIF5B and aIF1A, remains to be extensively addressed. Here, we study the late steps of Pyrococcus abyssi translation initiation. Using in vitro reconstituted initiation complexes and light scattering, we show that aIF5B bound to GTP accelerates subunit joining without the need for GTP hydrolysis. We report the crystallographic structures of aIF5B bound to GDP and GTP and analyze domain movements associated to these two nucleotide states. Finally, we present the cryo-EM structure of an initiation complex containing 30S bound to mRNA, Met-tRNAiMet, aIF5B and aIF1A at 2.7 Å resolution. Structural data shows how archaeal 5B and 1A factors cooperate to induce a conformation of the initiator tRNA favorable to subunit joining. Archaeal and eukaryotic features of late steps of translation initiation are discussed. |
 External links External links |  Nucleic Acids Res / Nucleic Acids Res /  PubMed:35694843 / PubMed:35694843 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 1.7 - 3.6 Å |
| Structure data | EMDB-14579, PDB-7zag: EMDB-14580, PDB-7zah: EMDB-14581, PDB-7zai: EMDB-14731, PDB-7zhg: EMDB-14763, PDB-7zki:  PDB-7yyp:  PDB-7yzn: |
| Chemicals |  ChemComp-GDP:  ChemComp-NO3:  ChemComp-HOH:  ChemComp-MG:  ChemComp-GTP:  ChemComp-NA:  ChemComp-ZN:  ChemComp-MET:  ChemComp-GNP: |
| Source |
|
 Keywords Keywords |  TRANSLATION / TRANSLATION /  ribosome / initiator tRNA / ribosome / initiator tRNA /  initiation factor / initiation factor /  eIF5B / IF2 / eIF5B / IF2 /  Initiation complex / Initiation complex /  translation initiation / small ribosomal subunit / aIF5b translation initiation / small ribosomal subunit / aIF5b |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers