+Search query
-Structure paper
| Title | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense. |
|---|---|
| Journal, issue, pages | Mol Cell, Vol. 83, Issue 24, Page 4586-44599.e5, Year 2023 |
| Publish date | Dec 21, 2023 |
 Authors Authors | Zhangfei Shen / Qingpeng Lin / Xiao-Yuan Yang / Elizabeth Fosuah / Tian-Min Fu /  |
| PubMed Abstract | SIR2-HerA, a bacterial two-protein anti-phage defense system, induces bacterial death by depleting NAD upon phage infection. Biochemical reconstitution of SIR2, HerA, and the SIR2-HerA complex ...SIR2-HerA, a bacterial two-protein anti-phage defense system, induces bacterial death by depleting NAD upon phage infection. Biochemical reconstitution of SIR2, HerA, and the SIR2-HerA complex reveals a dynamic assembly process. Unlike other ATPases, HerA can form various oligomers, ranging from dimers to nonamers. When assembled with SIR2, HerA forms a hexamer and converts SIR2 from a nuclease to an NAD hydrolase, representing an unexpected regulatory mechanism mediated by protein assembly. Furthermore, high concentrations of ATP can inhibit NAD hydrolysis by the SIR2-HerA complex. Cryo-EM structures of the SIR2-HerA complex reveal a giant supramolecular assembly up to 1 MDa, with SIR2 as a dodecamer and HerA as a hexamer, crucial for anti-phage defense. Unexpectedly, the HerA hexamer resembles a spiral staircase and exhibits helicase activities toward dual-forked DNA. Together, we reveal the supramolecular assembly of SIR2-HerA as a unique mechanism for switching enzymatic activities and bolstering anti-phage defense strategies. |
 External links External links |  Mol Cell / Mol Cell /  PubMed:38096827 PubMed:38096827 |
| Methods | EM (single particle) |
| Resolution | 2.83 - 3.6 Å |
| Structure data | EMDB-40762, PDB-8su9: EMDB-40763, PDB-8sub: EMDB-40778, PDB-8suw: EMDB-40860, PDB-8sxx: EMDB-42061, PDB-8uae: EMDB-42062, PDB-8uaf: |
| Chemicals | 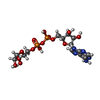 ChemComp-AR6:  ChemComp-ADP:  ChemComp-MG:  ChemComp-NAD:  ChemComp-AGS: |
| Source |
|
 Keywords Keywords |  IMMUNE SYSTEM / HerA / IMMUNE SYSTEM / HerA /  SIR2 / NADase / SIR2 / NADase /  ATPase / Anti-phage system / Anti-phage / ATPase / Anti-phage system / Anti-phage /  Nuclease Nuclease |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers
















